01 - BLAST+ at NCBI¶
Learning Outcomes¶
- Use of
BLAST+at theNCBIwebsite, using the browser interface - Obtaining and downloading
BLAST+output in multiple formats - Interpreting
BLAST+output
Table of Contents¶
Introduction¶
BLAST can be run using any of several interfaces, including:
- at the NCBI website: https://blast.ncbi.nlm.nih.gov/Blast.cgi
- using the
ncbi-blastprogram suite in a terminal - by interacting with the tool, using a programming language
- creating a new instance in the cloud, to run your own
BLASTserver - sending a job to the NCBI servers, using a programming language
In this part of the course, we will use the first three methods to explore ways of using BLAST. In this markdown notebook, we will use BLAST at the NCBI website, but in the next two notebooks we will use a local installation of BLAST on your laptop or a Linux virtual machine (VM).
Why so many ways to use BLAST?¶
- The web interface at NCBI is user-friendly, and available wherever you have a network connection. It uses the large computing resources of NCBI to run queries against very large databases very quickly. A number of variants to the standard
BLASTsearch are offered, which may be appropriate for a particular task. - Using a local installation of
ncbi-blastat the terminal gives you full control over how to use the program, and allows you to readily build custom databases (useful for proprietary information). However, you are limited to the computing power you have available. Happily,BLASTdoesn't require excessive amounts of computing resources and for many tasks a desktop or laptop machine is sufficient. - Programmatic running of
BLAST- either locally or using the web interface to the NCBI servers - allows for repeated searches, and automated integration of the search results with arbitrary data-processing tasks and other analyses.
Using the NCBI website¶
- Open a web browser and navigate to the NCBI
BLASTwebpage.
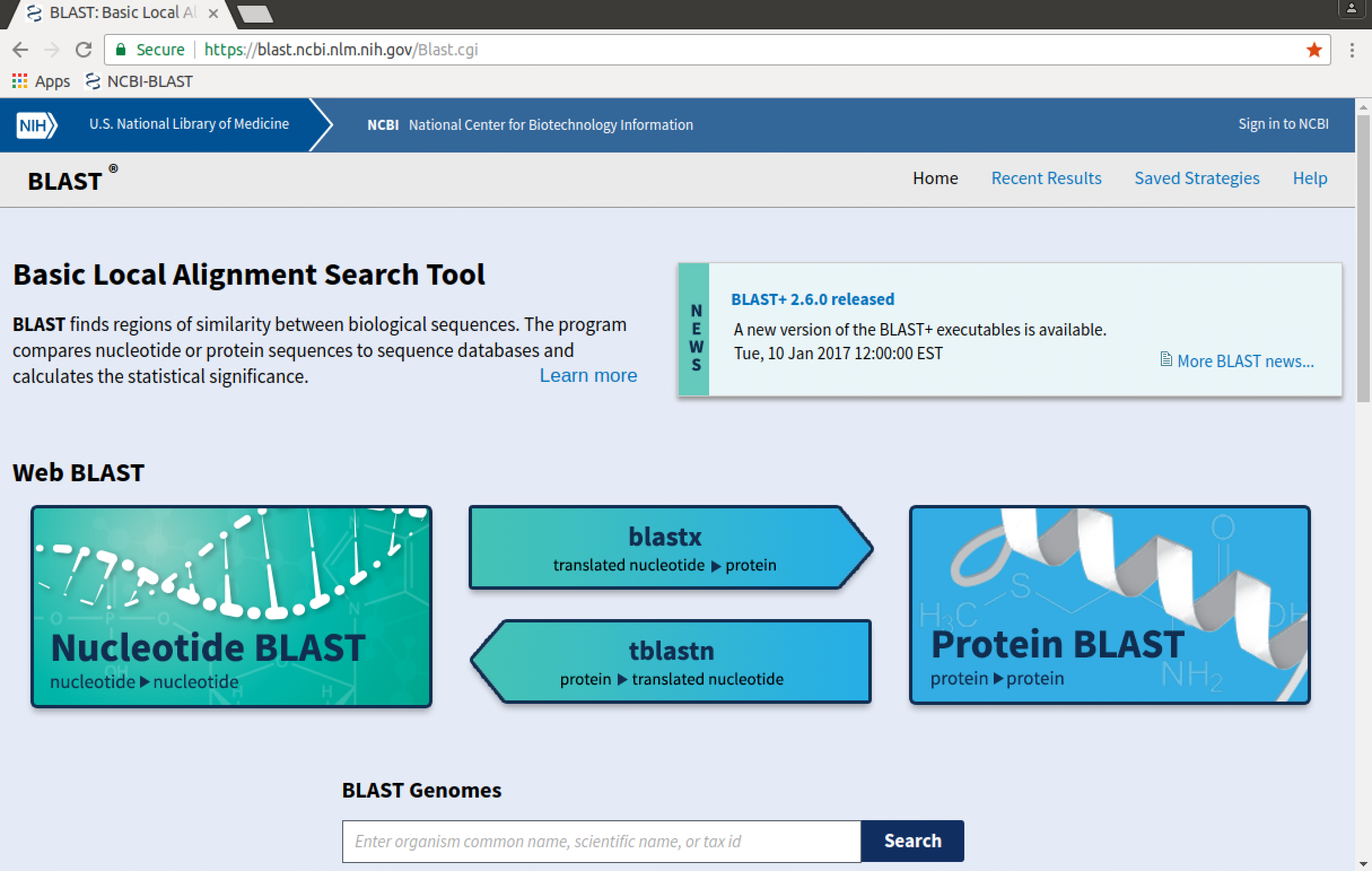
Scrolling up-and-down the page indicates several available BLAST tools, including:
- 'Nucleotide BLAST' (
BLASTN- query a nucleotide sequence against a database of nucleotide sequences) - 'Protein BLAST' (
BLASTP- query a protein sequence against a database of protein sequences) BLASTX(query a nucleotide sequence against a database of protein sequences)TBLASTN(query a protein sequence against a database of nucleotide sequences)- SmartBLAST (query a protein against the specialised "landmark" database to find highly-similar sequences)
- Primer-BLAST (find primers specific to your PCR template)
- CD-search (search specifically for conserved domains of a sequence)
- GEO (query a nucleotide sequence against a database of transcriptional expression profiles)
- Targeted Loci (query a nucleotide sequence against a database of sequence markers)
- Click on the "Nucleotide BLAST" link to get the web
BLASTNinterface.
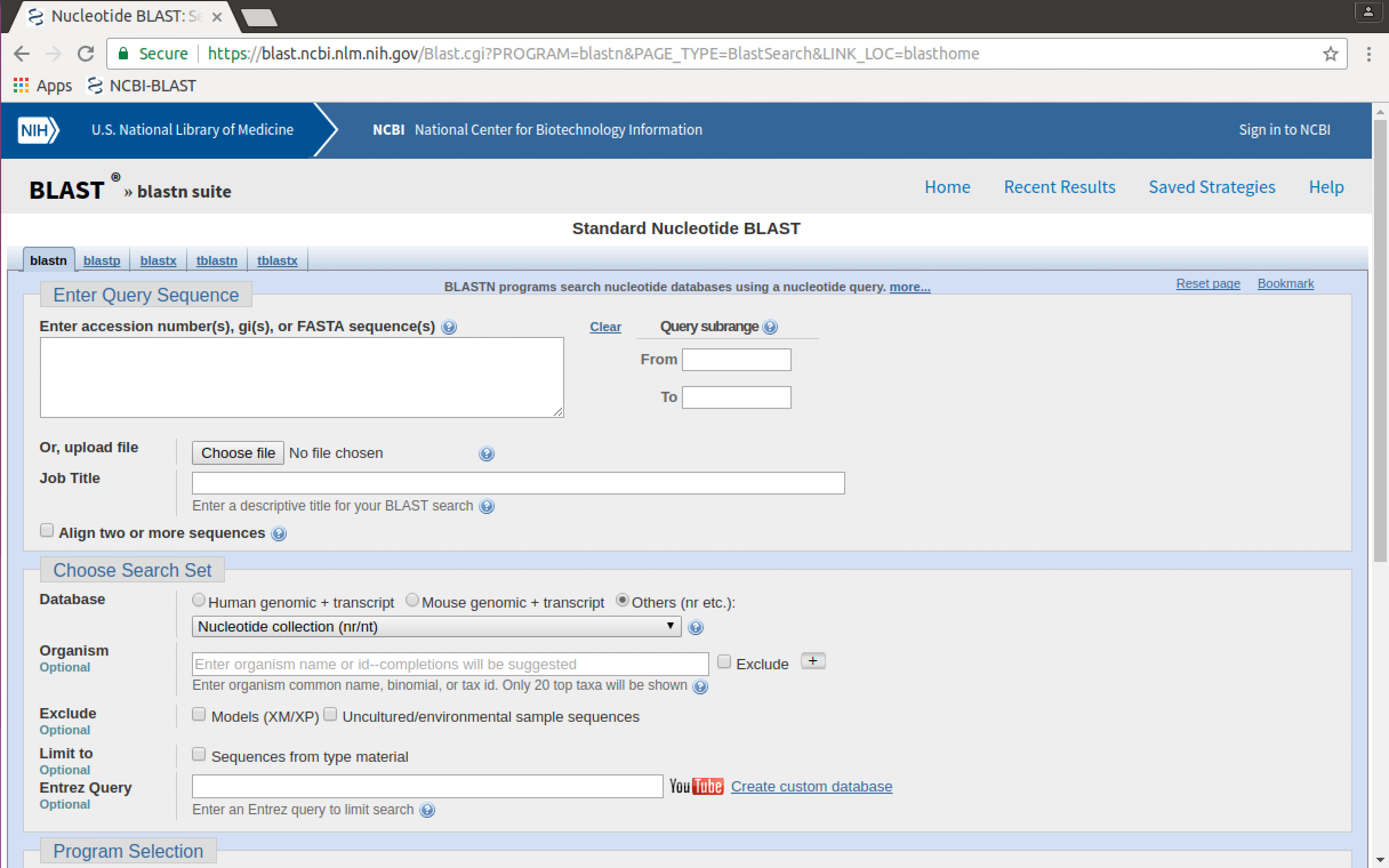
Sequences can be typed or copied and pasted into the "Enter Query Sequence" box, but they can also be uploaded from an existing FASTA file on your computer. We will do this.
- Click on the "Choose file" button. A file dialogue will pop up.
- In the file dialogue, navigate to, and select the file
k_sp_CB01950_penicillin.fastain the02-sequence_databases/data/kitasatosporadirectory. Then click on "Open".
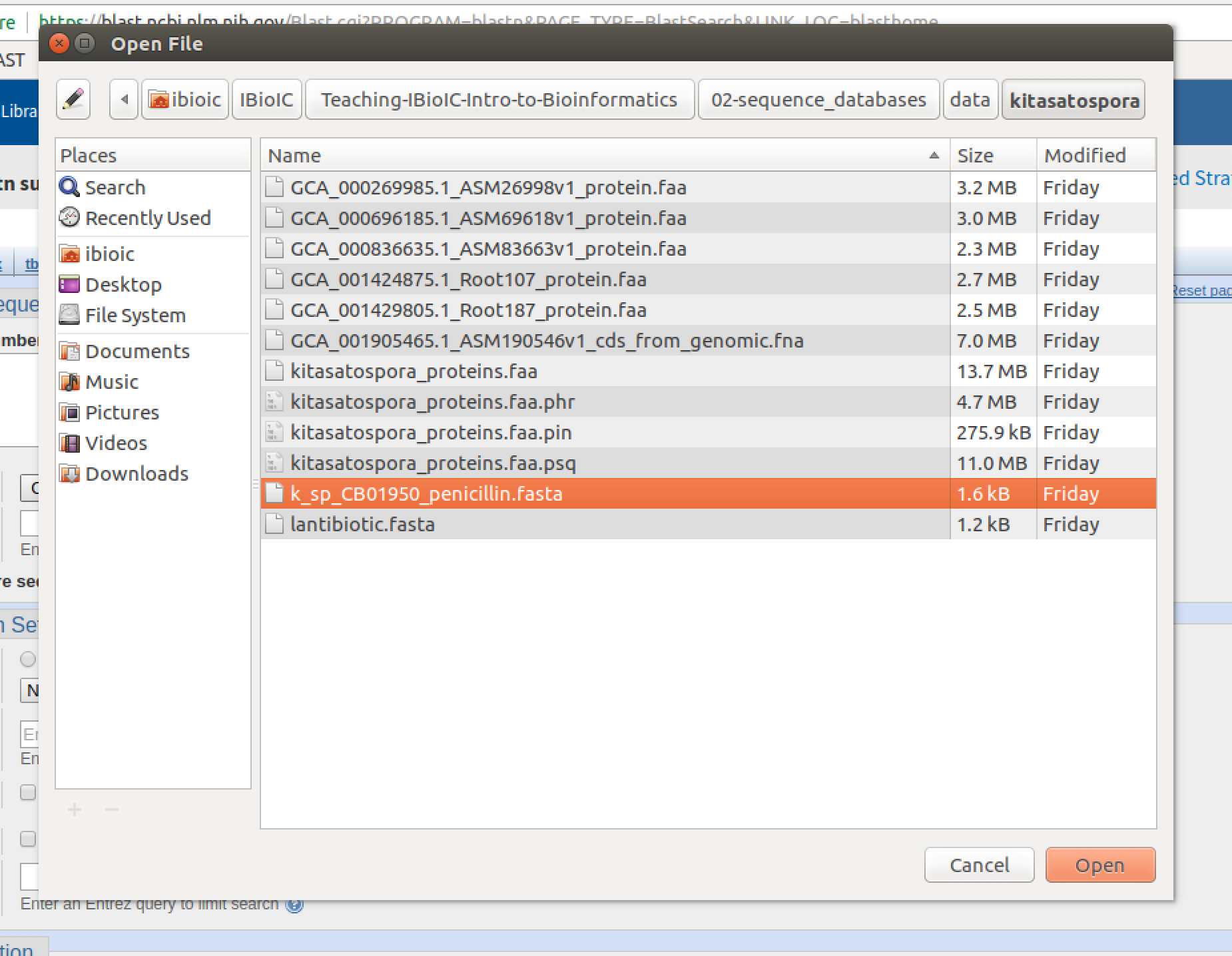
The file will show up as being selected next to the "Choose file" button.
- Enter a descriptive job title in the Job Title field
- Make sure the selected database is "Nucleotide collection"
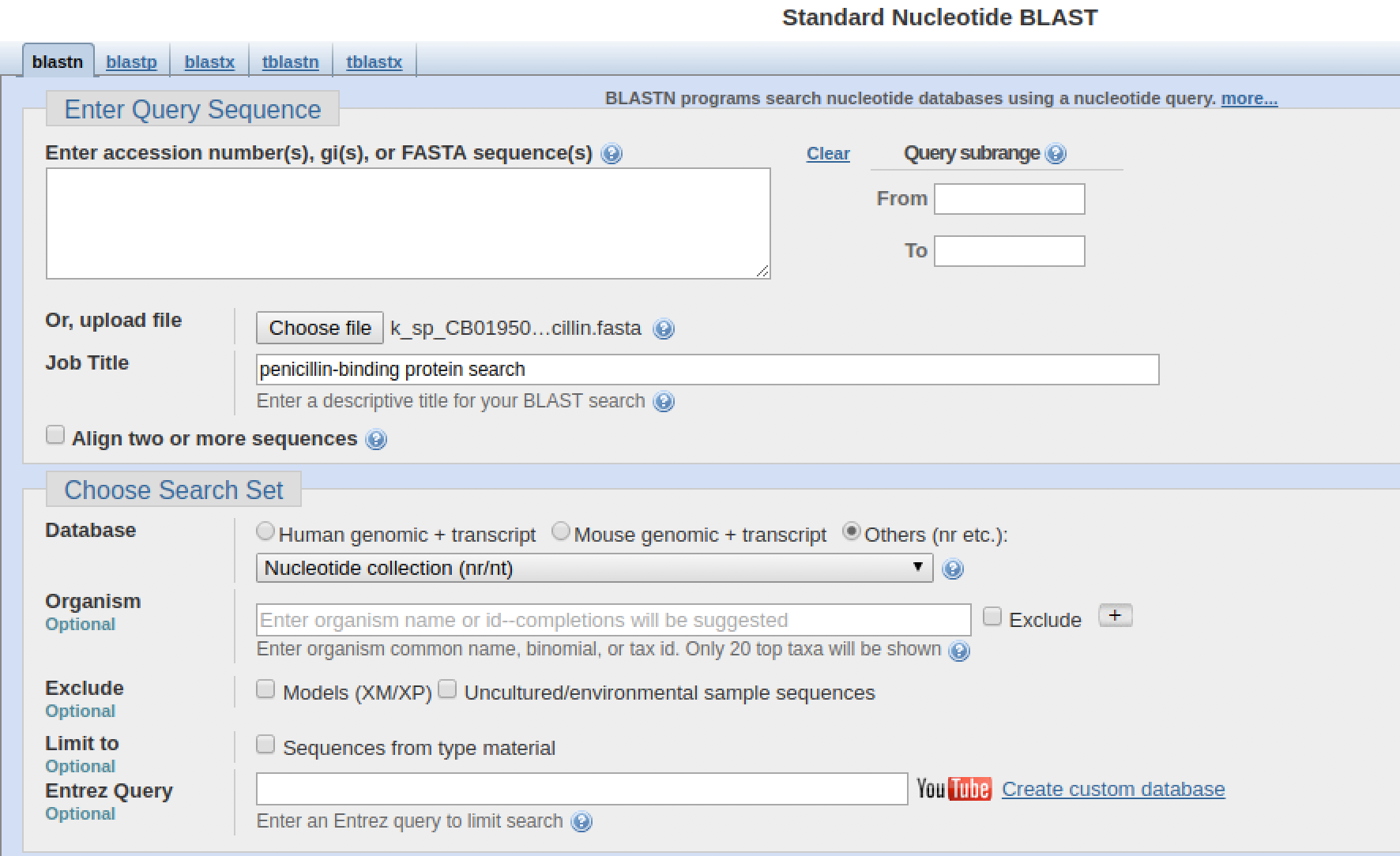
- Scroll to the bottom of the page and click on the "BLAST" button.
An interstitial page will appear, reporting the job ID, and giving you runtime information. The page will be updated automatically, and frequently.
Once the job is complete, the results will be displayed as an interactive webpage.
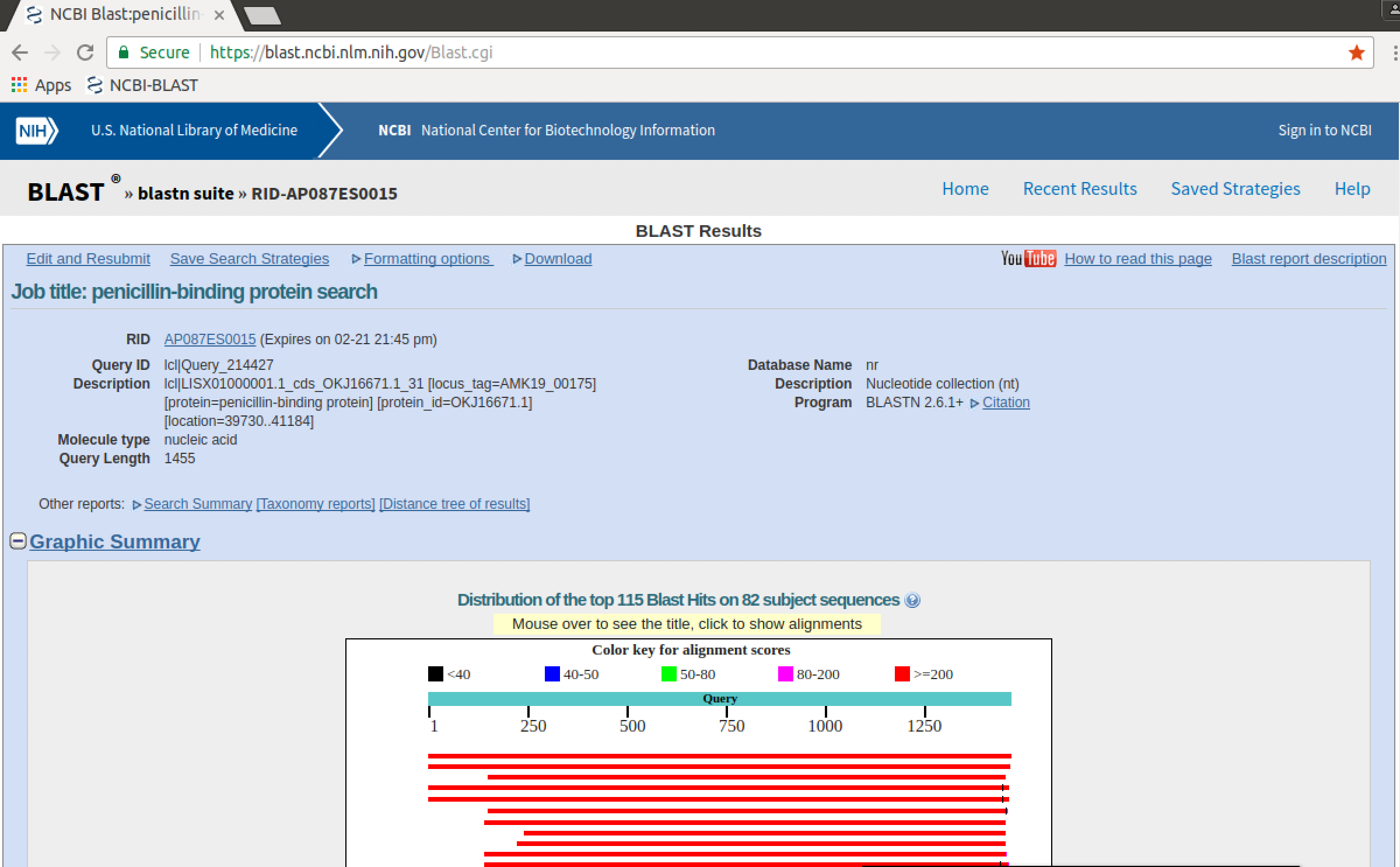
- Inspect the results - note the information that the page gives you about how the search was done.
QUESTIONS
- What is the "best hit" to the query? Why do you think it is the "best hit" (what in the results tells you this?)
- At what point do you think the matches start to become less reliable? Why do you think this? (*HINT:* inspect the alignments)
- Click on the links to `[Taxonomy reports]` and `[Distance tree of results]` at the top of the page. What information do these reports add to the main result?
Download the results¶
- Click on the
Downloadlink at the top of the results page.
- Right-click on the download link for
Textand save the results tooutput/kitasatospora/ncbi_blastn_query_01.txt - Right-click on the download link for
Table(CSV)and save the results tooutput/kitasatospora/ncbi_blastn_query_01.csv
Exercise 01: Using the NCBI BLAST Website¶
Using the NCBI BLAST website:
- Conduct a `BLASTX` query with `data/kitastaospora/lantibiotic.fasta` against the `nr` database, restricting your results only to *Kitasatospora* spp. matches (taxid: 2063)
- Save the results in `Text` and `Table(CSV)` format to
- `output/kitasatospora/ncbi_blastx_query_02.txt`
- `output/kitasatospora/ncbi_blastx_query_02.csv`
QUESTIONS
- How many hits do you find?
- What is the "best hit" to the query? Why do you think it is the "best hit" (what in the results tells you this?)
- At what point do you think the matches start to become less reliable? Why do you think this? (*HINT:* inspect the alignments)