02-BLAST+ at the terminal¶
Learning Outcomes¶
- Use of
BLAST+at the terminal - Getting help at the command line
- Building custom local
BLAST+databases - Understanding the
BLAST+command-line - Getting output in multiple formats
- Interpreting
BLAST+output
Introduction¶
The BLAST/BLAST+ package can be installed on your own machine (desktop or laptop) or on a shared server. This gives you full control over how to use the program, and allows you to build custom databases (useful for proprietary information). However, you are limited to the computing power you have available. Happily, BLAST doesn't require excessive amounts of computing resources and for many tasks a desktop or laptop machine is sufficient.
Using BLAST+ in the terminal¶
- If necessary, open a terminal window in the virtual machine (VM), or a new tab in your terminal window.
- Change directory to the
~/Teaching-IBioIC-Intro-to-Bioinformatics/02-sequence_databaseslesson directory:
cd ~/Teaching-IBioIC-Intro-to-Bioinformatics/02-sequence-databases
ls
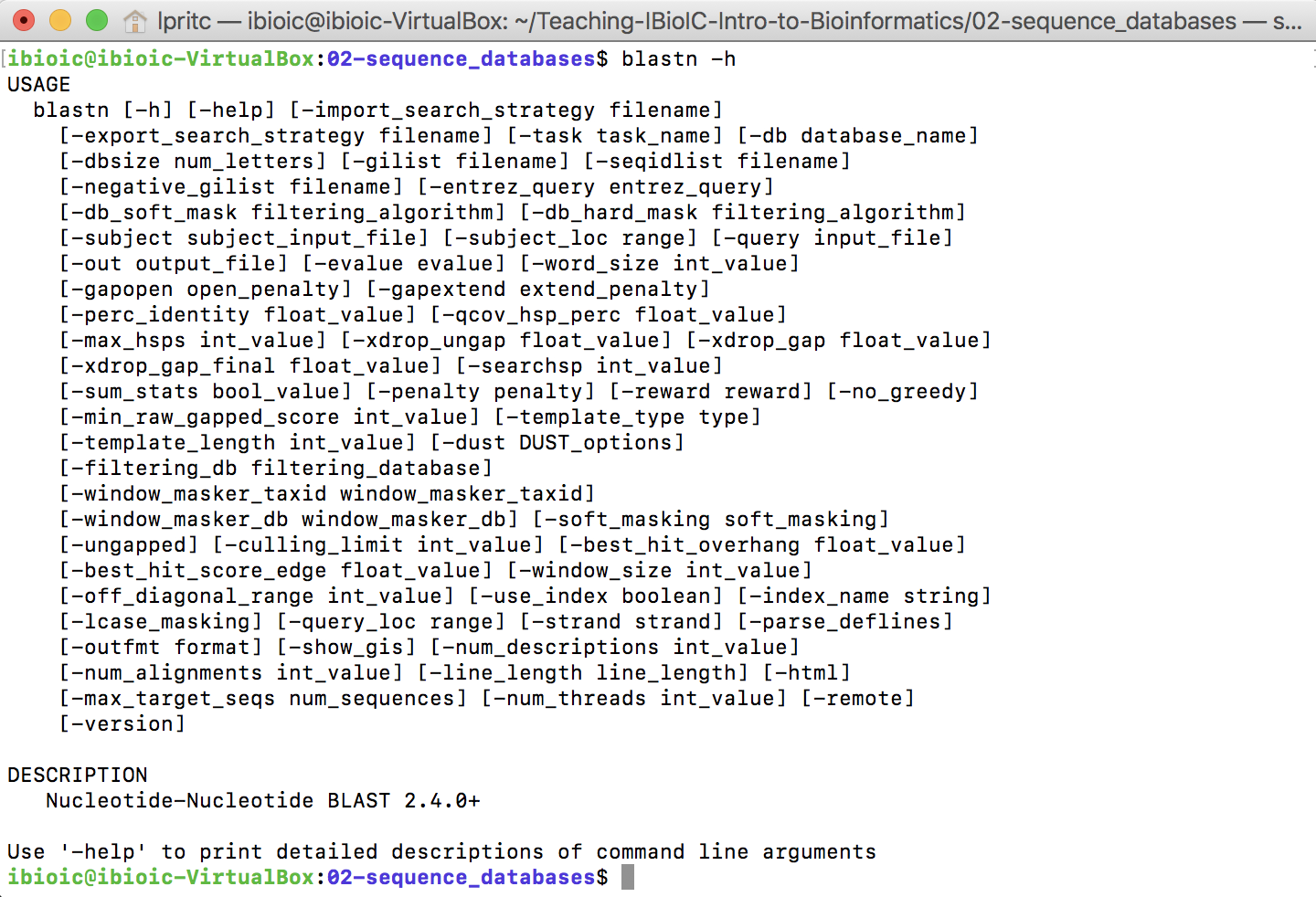
- Establish that
BLASTNworks by issuing a command to get the short help message:
blastn -h
Build a BLAST+ database¶
The program that builds BLAST+ sequence databases is makeblastdb. You can get basic help on the command by issuing:
makeblastdb -h
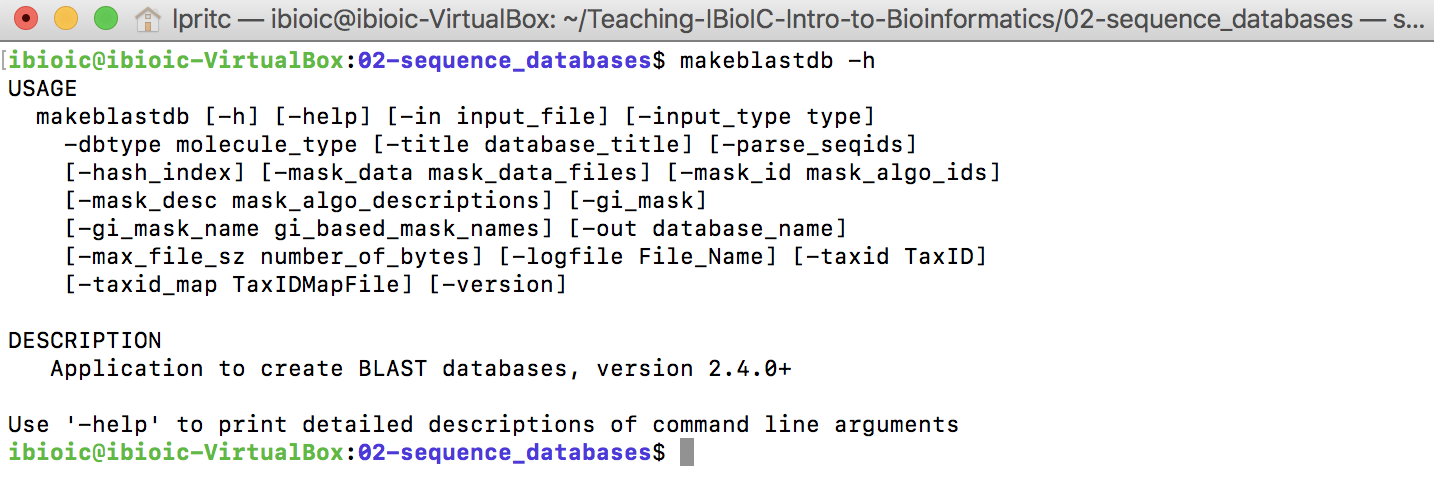
- A file containing the sequences that will be in the database
- What kind of sequence (nucleotide or protein) data the file contains
- A name for the database (optional)
- A path to write the database files to (optional)
- Create a new
BLASTdatabase with the following command:
makeblastdb -in data/kitasatospora/GCA_001905465.1_ASM190546v1_cds_from_genomic.fna \
-dbtype nucl \
-title kitasatospora_cds \
-out data/kitasatospora/kitasatospora_cds
This will return some information to the terminal, and create the database.
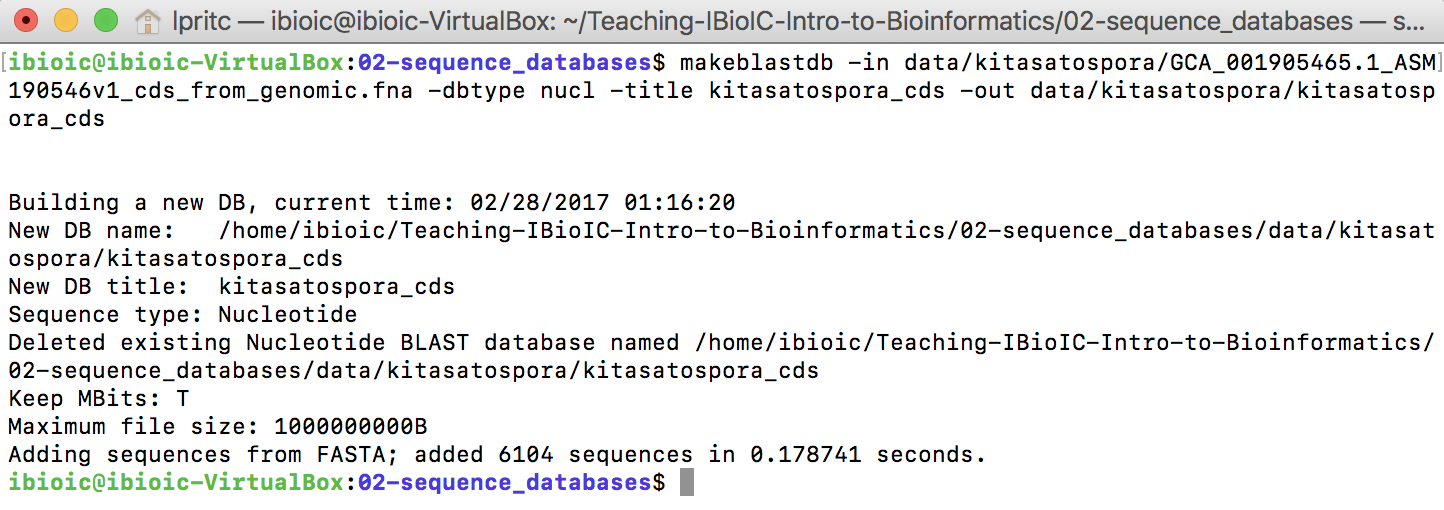
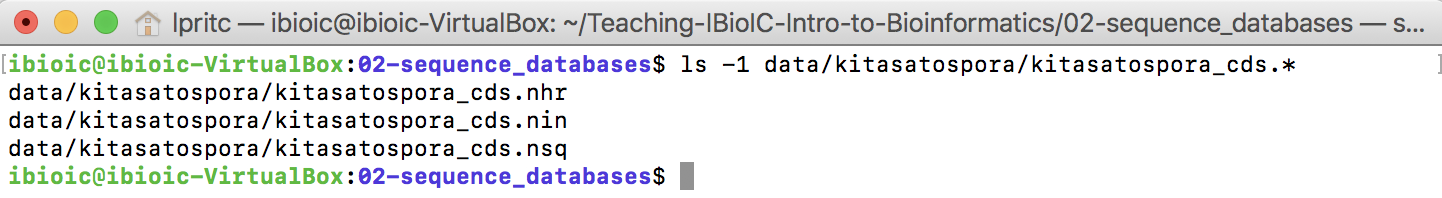
This creates three files, which together comprise a new BLAST nucleotide database against which you can make queries.
Exercise 01: Get BLAST help at the Terminal¶
Use the following commands to get the long-format help messages for BLASTN and BLASTX:
- `blastn -help`
- `blastx -help`
Construct a BLASTN query¶
After looking at the help information in the exercise above, you will have seen that there are several relevant input options:
-query: path to the query sequence(s)-db: path to theBLASTdatabase-outfmt: the output format you wantBLASTto produce-out: path to the output file you wantBLASTto write
In this case:
- your query sequence is
data/kitasatospora/lantibiotic.fasta - the database you're searching against is the one you created above:
data/kitasatospora/kitasatospora_cds
and you'll generate output in two formats (the same ones that were produced from the NCBI website search). You will need to construct two commands, each with the same query and database, but different output format values, and output filenames:
- no format specified, filename:
output/kitasatospora/terminal_blastn_query_01.txt - format:
6(tabular), filename:output/kitasatospora/terminal_blastn_query_01.tab - Run the first command at the terminal:
blastn -query data/kitasatospora/lantibiotic.fasta \
-db data/kitasatospora/kitasatospora_cds \
-out output/kitasatospora/terminal_blastn_query_01.txt
head -n 40 output/kitasatospora/terminal_blastn_query_01.txt
- Run the second command, now specifying a different (tabular) output format:
blastn -query data/kitasatospora/lantibiotic.fasta \
-db data/kitasatospora/kitasatospora_cds \
-outfmt 6 \
-out output/kitasatospora/terminal_blastn_query_01.tab
You can inspect the contents of this file by issuing the command:
less output/kitasatospora/terminal_blastn_query_01.tab
QUESTIONS
- How many hits were found
- How large was the database?
- How does the tabular output compare to the plain text output?
Exercise 02: Using BLAST at the Terminal¶
Using BLAST in the terminal:
- Conduct a `BLASTX` query with `data/kitastaospora/lantibiotic.fasta` against the `data/kitasatospora/kitasatospora_proteins.faa` database, writing results in `Text` and `Tabular` format to:
- `output/kitasatospora/terminal_blastx_query_02.txt`
- `output/kitasatospora/terminal_blastx_query_02.tab`
QUESTIONS
- How many hits do you find?
- What is the "best hit" to the query? Why do you think it is the "best hit" (what in the results tells you this?)
- At what point do you think the matches start to become less reliable? Why do you think this? (*HINT:* inspect the alignments)