06 - UniProt¶
Table of Contents¶
Introduction¶
UniProt provides three key databases:
- The UniProt Knowledgebase (UniProtKB)
- The UniProt Reference Clusters (UniRef)
- The UniProt Archive (UniParc)
UniProtKB is likely to be the database you use most frequently to find information on gene product/protein molecular function. It is the central hub of functional information on proteins, and collates functional annotations from many other databases, ontologies, and references. It keeps records of how annotations are derived (e.g. experimentally or computationally), and is divided into two sections: one contains manually-annotated records (UniProtKB/Swiss-Prot), and a second contains computational annotations that are waiting for manual curation (UniProtKB/TrEMBL).
UniParc is a comprehensive, non-redundant database that contains most of the publicly-available protein sequences from a range of sources.
UniRef provides clustered sets of sequences from UniProtKB and UniParc. A number of clusterings at different stringencies are provided.
These databases can be queried in a number of ways, including:
- At the
UniProtwebsite http://www.uniprot.org/ in your web browser - Sending requests to the
UniProtwebsite, using a programming language
Resources¶
The UniProt website¶
- Open a web browser and navigate to the
UniProtwebpage.
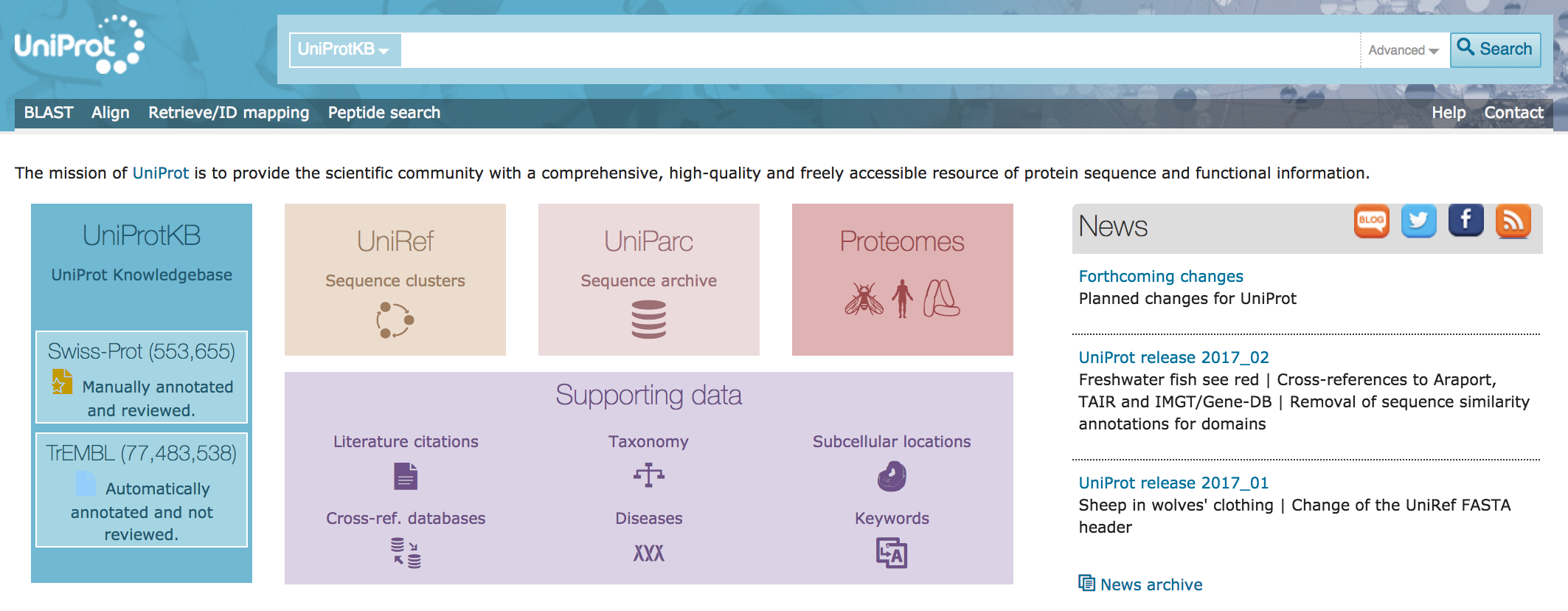
The landing page offers options for each of the three main databases: UniProtKB, UniRef, and UniParc. It also offers sets of complete proteomes for a range of organisms, and databases of proteins organised by supporting data, such as literature, taxonomic classification, and subcellular location.
Using UniProtKB¶
- Click on the
UniProtKBlink. This will take you to the UniProtKB front page, with a summary of entries, and a number of links.
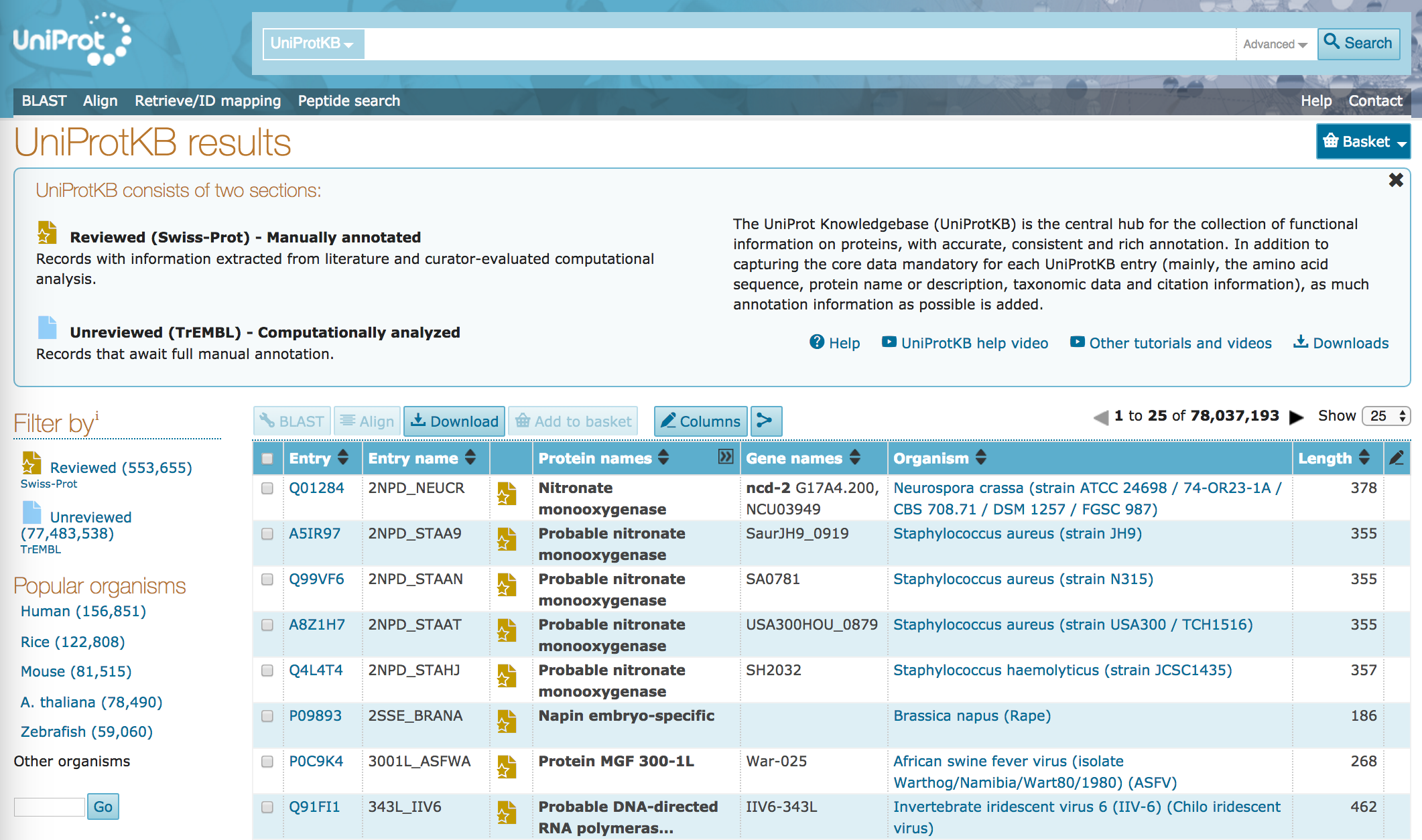
- How many records are in UniProtKB today?
- How many of those records have been manually reviewed? What proportion of the total database has been manually reviewed?
- Which organisms are most highly represented in the database, today?
- Enter the word Kitasatospora in the search bar at the top of the page, and click on the
Searchbutton.

- How many entries are returned?
- How many of those entries have been manually reviewed? What proportion of the total is this?
Filtering Results¶
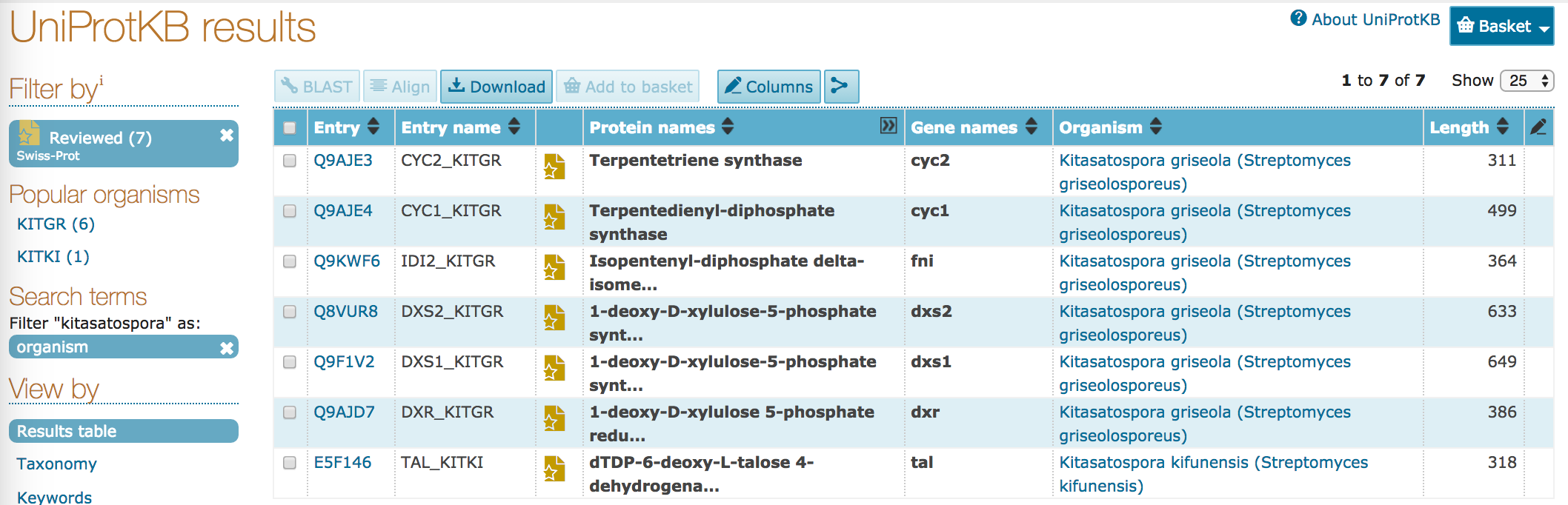
- On the left of the page there's an option to filter
"kitasatospora"as an organism or by taxonomy. Click on theorganismfilter.
- How many entries are returned?
- How many of those entries have been manually reviewed?
- How has the search term in the top bar changed? NOTE: these search term changes will be useful for querying UniProt programmatically.
- At the top left of the page there's an option to filter only the manually reviewed entries. Click on this filter.
Inspecting an Entry¶
- Click on the link/accession for the topmost entry.
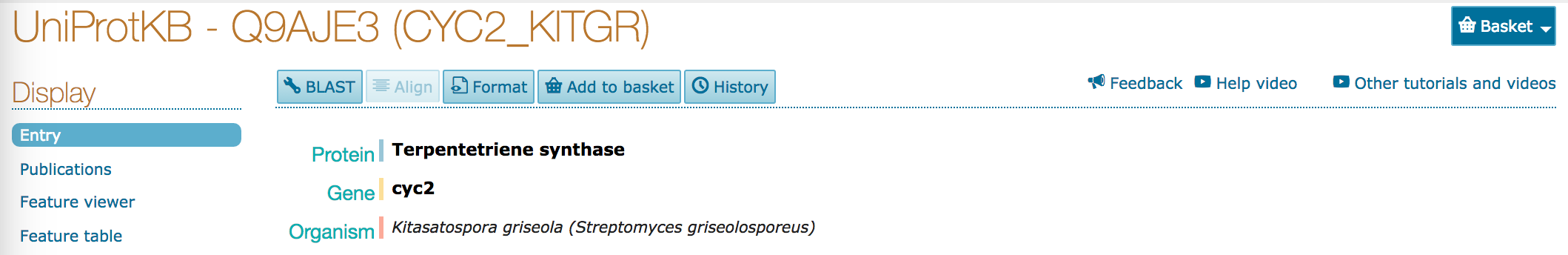
- What kind of evidence is there for protein function?
- Kinetic information for this enzyme is drawn from which other database(s)?
- Are any protein structures available for this enzyme?
- At the top of the page, there's a button marked
History. Click on this button. A small window will open, with a link toPrevious versions. Click on this link.
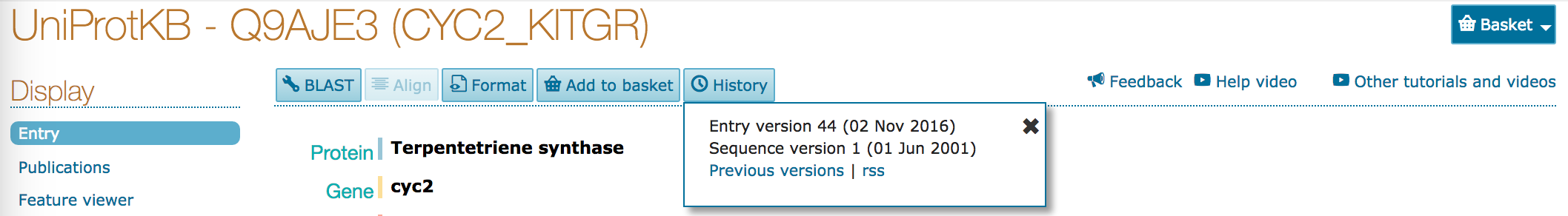
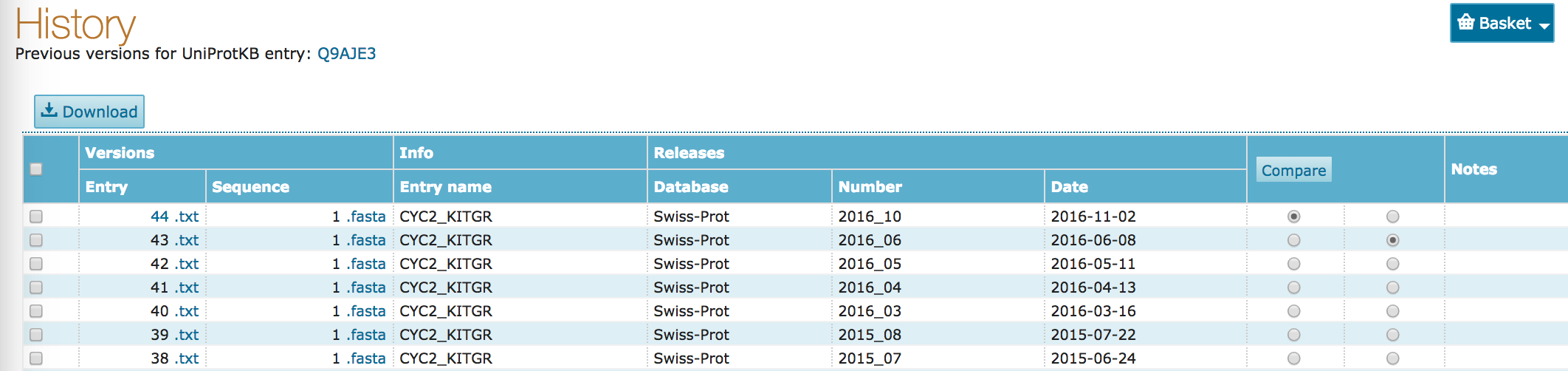
- Click on the
Comparebutton.
- When was the last change made to this record?
- What was the change?
- Compare some previous records to the current record (e.g. this comparison).
- What kinds of changes do you see?
Advanced Searches in UniProtKB¶
In this section, you'll use the advanced searches to identify candidate human proteins that are found in the nucleus, and have been associated with some disease activity or function.
- Click on the UniProt logo to return to the landing page
- Click on the "Advanced" drop-down to get the advanced searching interface
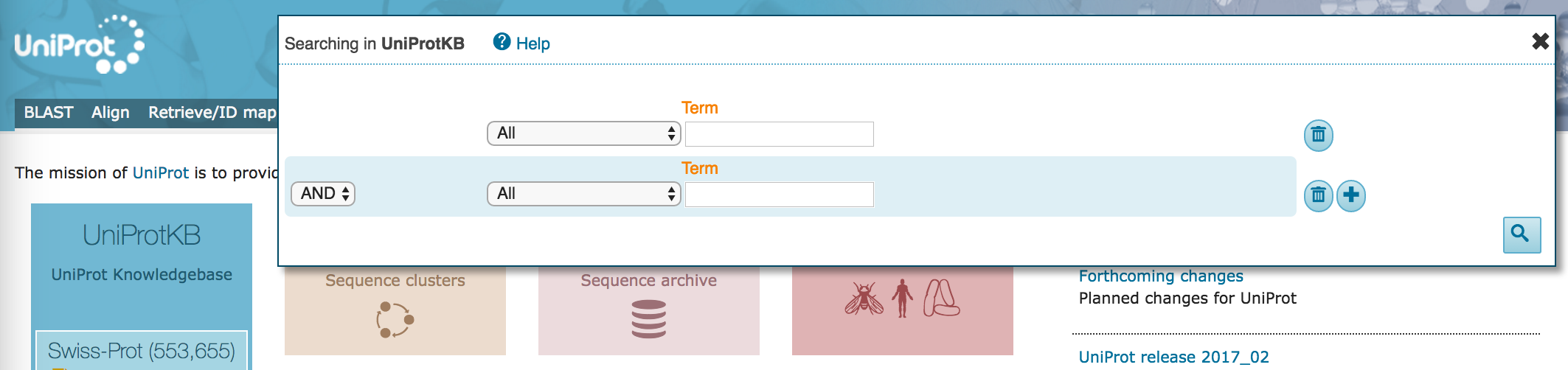
- In the first field, select
Organism [OS]with search term "Human". The dropdown will offer you several options as you type, but do not select them (you could have entered the organism "Homo sapiens" here, also). - In the next field, keep
ANDon the left, and selectSubcellular locationwith search term "nucleus". The dropdown will offer several options but, again, do not select them. At this point, allow any assertion method for the evidence code.
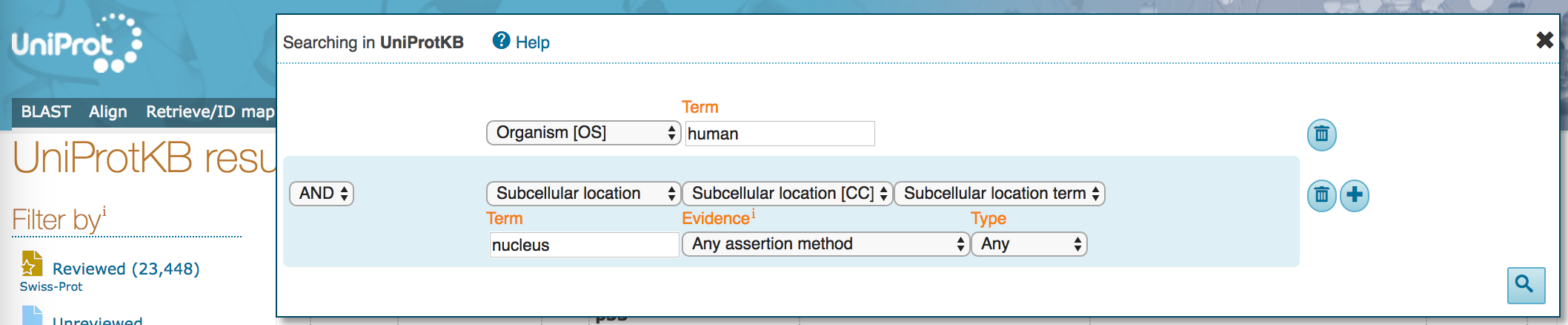
- Click on the plus sign (
+) to get another search term field. - In the new field, keep
ANDon the left, and selectPathology & Biotechwith class "Disease", and no search term. At this point, allow any assertion method for the evidence code.
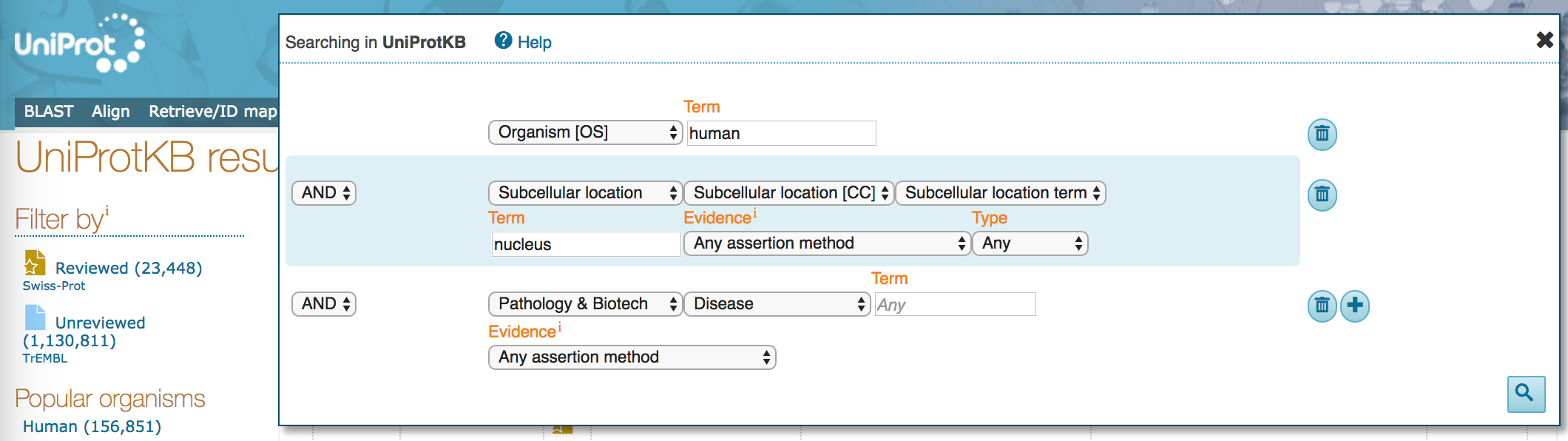
- Click on the magnifying glass to run the search.
- How many results do you see, today?
- What are the contents of the search bar? **NOTE: this will be useful for programmatic queries, later.**
Refine the search¶
- Click on the "Advanced" drop down. You should see that the current search populates this dialogue box.
- Change the
Evidenceoption for thePathology & Biotechpart of the search to manual "Experimental" evidence.
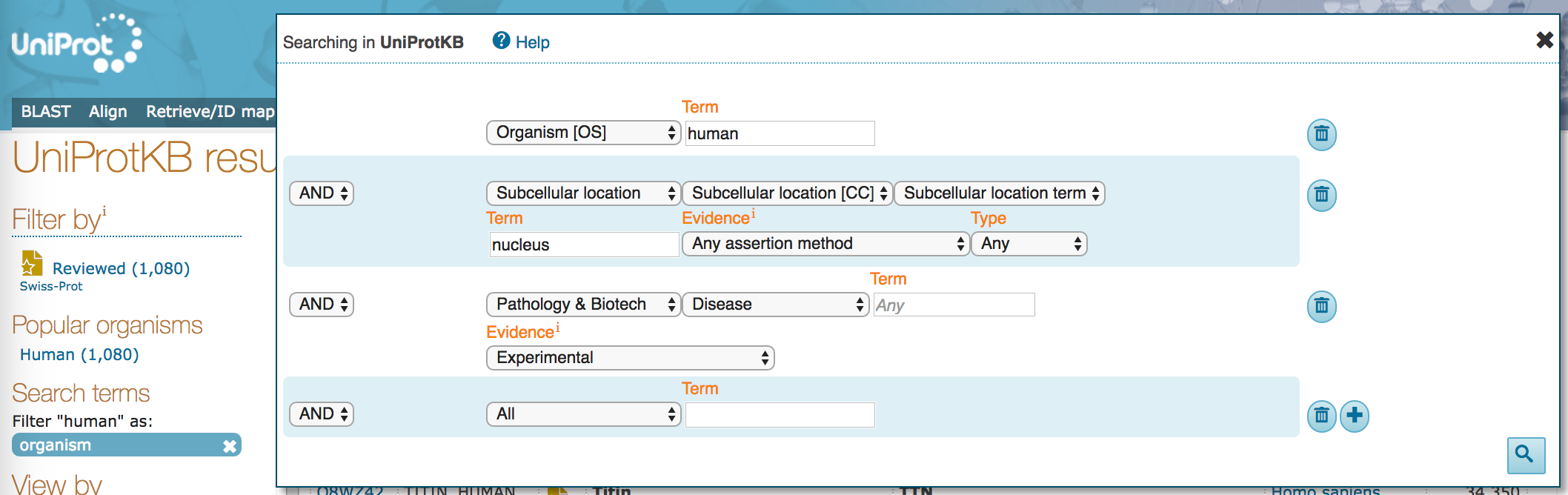
- How many results do you see, now?
- What are the contents of the search bar? **NOTE: this will be useful for programmatic queries, later.**
- Click on the "Advanced" drop down. You should see that the current search populates this dialogue box.
- Change the
Termoption for thePathology & Biotechpart of the search to "melanoma".
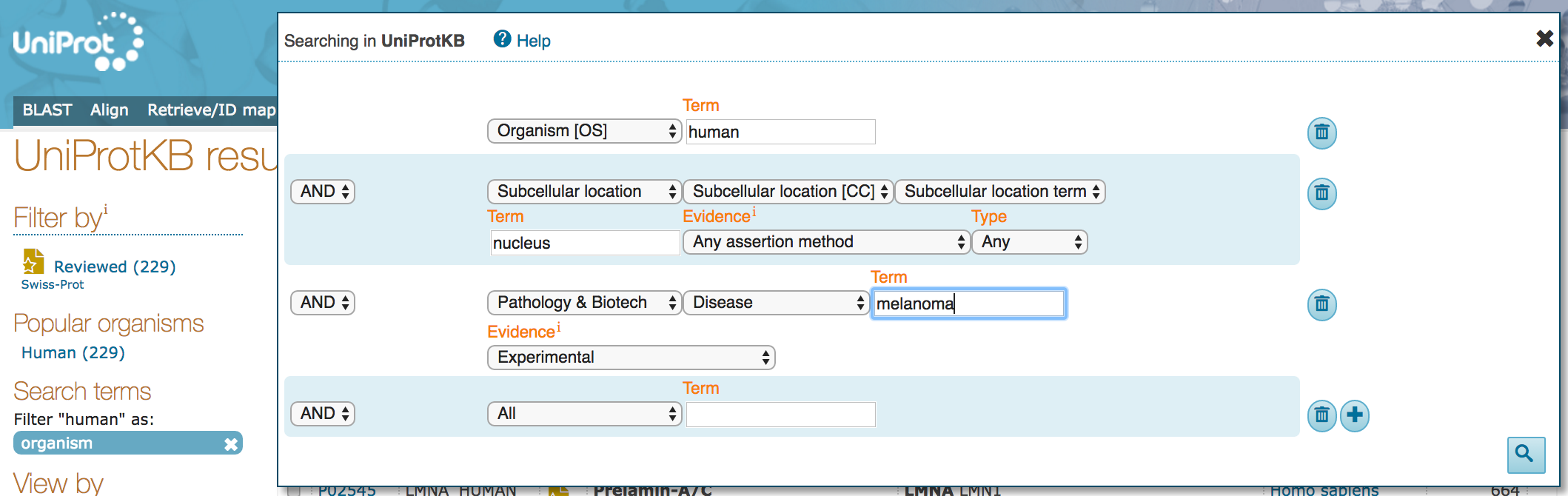
- How many results do you see, now?
- How are these proteins associated with melanoma?
- What amino acid modifications have been found for these proteins?
Downloading UniProtKB Search Results¶
After the search above, you should be left with a small set of proteins that satisfy the following criteria:
- They derive from Homo sapiens
- They are annotated as being found in the nucleus (for which we allow any form of evidence)
- They are associated with a disease process: melanoma, and there is manually-curated experimental evidence for this association
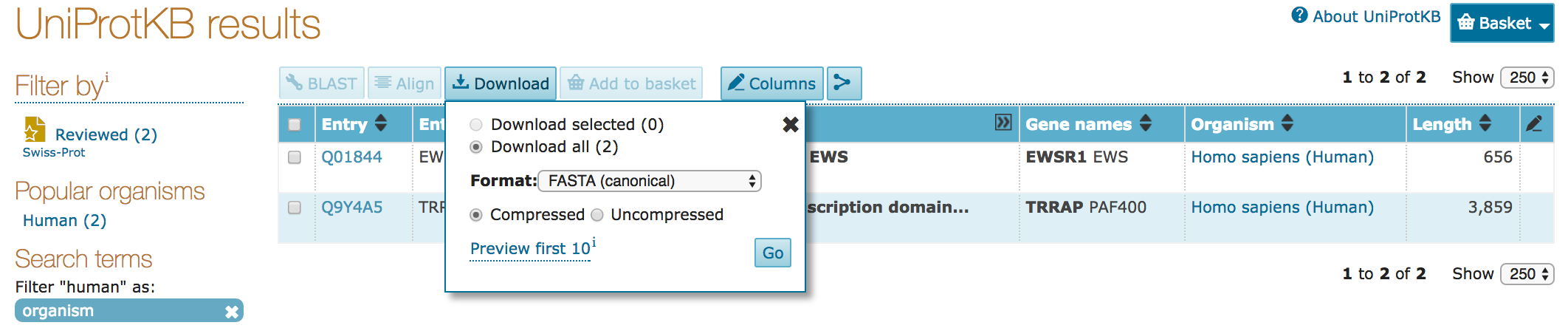
- You can download all your search results, or just those selected with checkboxes
- Results can be downloaded compressed (
gzipped) or as raw records - Results can be downloaded as:
- sequence data (
FASTA) - tabular form (
Excel,tab-separated) - computer-readable (
XML,RDF)
- sequence data (
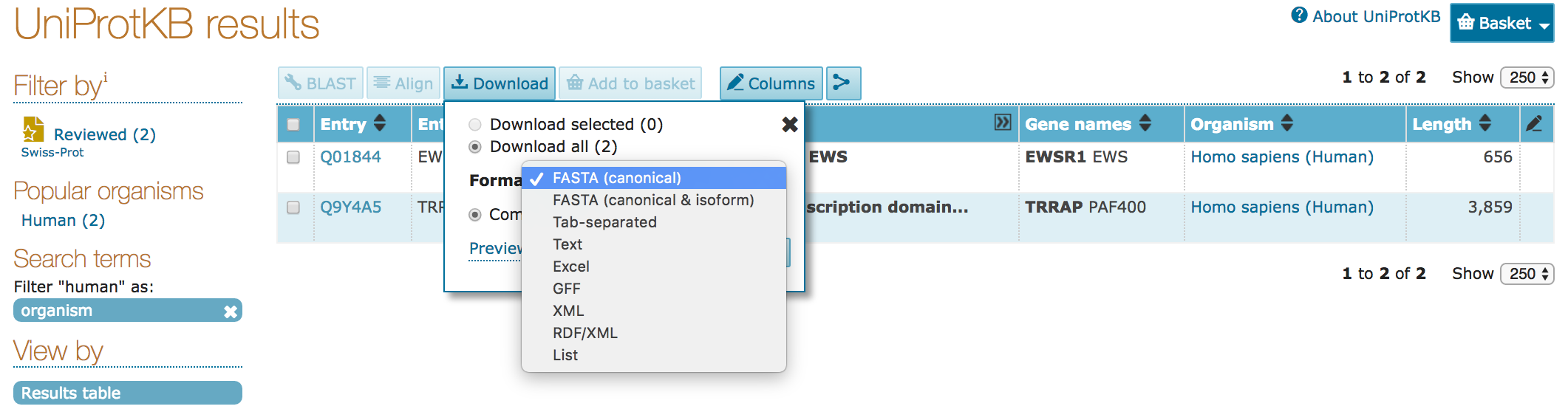
- Download the search results as tab-separated, text, and FASTA format files
- Inspect the contents of these files
- How do the contents of these files differ
Exercise 01 (15min)¶
Using the UniProtKB search tools, can you find and download sets of proteins that satisfy the following requirements:
Set 1
- Derives from *Saccharomyces cerevisiae*
- Is associated with a membrane
- Is a transcriptional regulator
Set 2
- Has a function associated with alginate
- Is annotated with any biotechnological application
Set 3
- Any manual annotation associated with biofuel as a biotechnological application
Set 4
- Derives from mouse
- Is an enzyme
- Has not been manually reviewed
- Is mentioned in a *Nature* publication
- Is between 100 and 300aa in length