08 - KEGG¶
Table of Contents¶
Introduction¶
The pathway maps - 'wiring diagrams' of molecular interconversions - are hand-drawn, and form the core of KEGG. They integrate many molecular types, including proteins, RNA, primary and secondary metabolites, and chemical reactions. The maps are classified in a number of ways (metabolism, disease, and so on), each of which can be considered a conceptual 'view' onto the pathway data.
KEGG can be queried in a number of ways, including:
- At the
KEGGwebsite [http://www.genome.jp/kegg/pathway.html) in your web browser - Sending requests to the
KEGGwebsite, using a programming language
Resources¶
The KEGG website¶
- Open your web browser and navigate to the
KEGGwebpage.
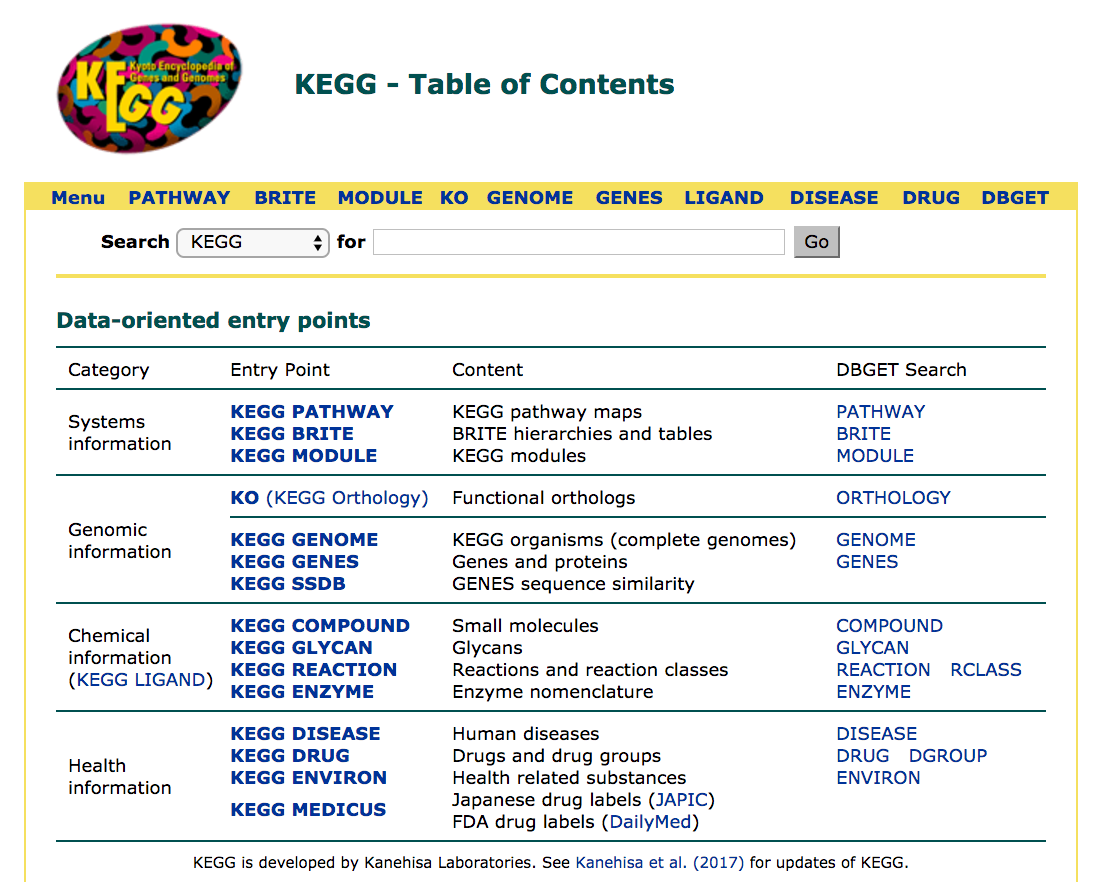
The landing page offers a menu of choices: PATHWAY, BRITE, MODULE and so on (the main page shown is the PATHWAY page) , which each link to different databases within KEGG. For example:
BRITE: Functional hierarchies and binary relationships of biological entitiesMODULE: Functional units for annotating and interpreting genomesKO: Linking genomes to pathways by ortholog annotation
A full account of all the databases at KEGG is beyond the scope of this lesson.
KEGG GENOME¶
KEGGGENOME: Organisms and ecosystems with genome sequence information
- Click on the GENOME link in the KEGG menu bar. This will take you to the
KEGGGENOME database landing page.
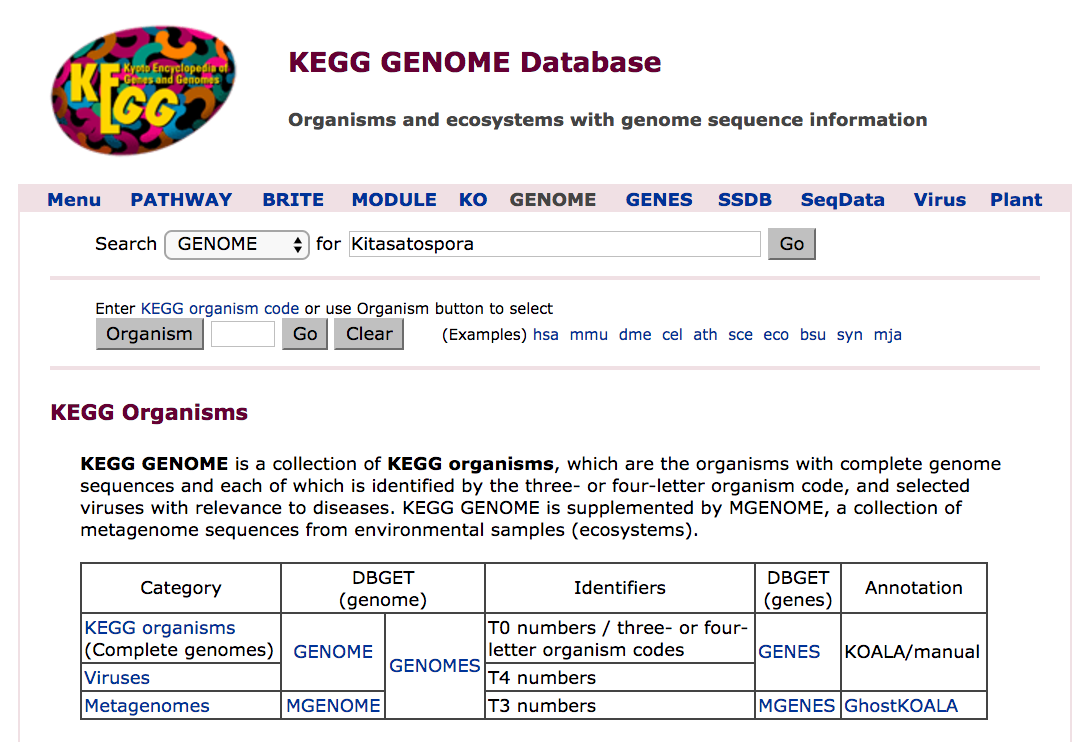
The landing page presents you with a search field.
- Enter the word "Kitasatospora" into the search field, and click on "Go"
- How many entries are returned?
- What is the theoretical upper limit on the number of three-letter codes? What does this imply about the capacity of `KEGG` GENOME's naming scheme?
- Click on the first link in the list of genomes.
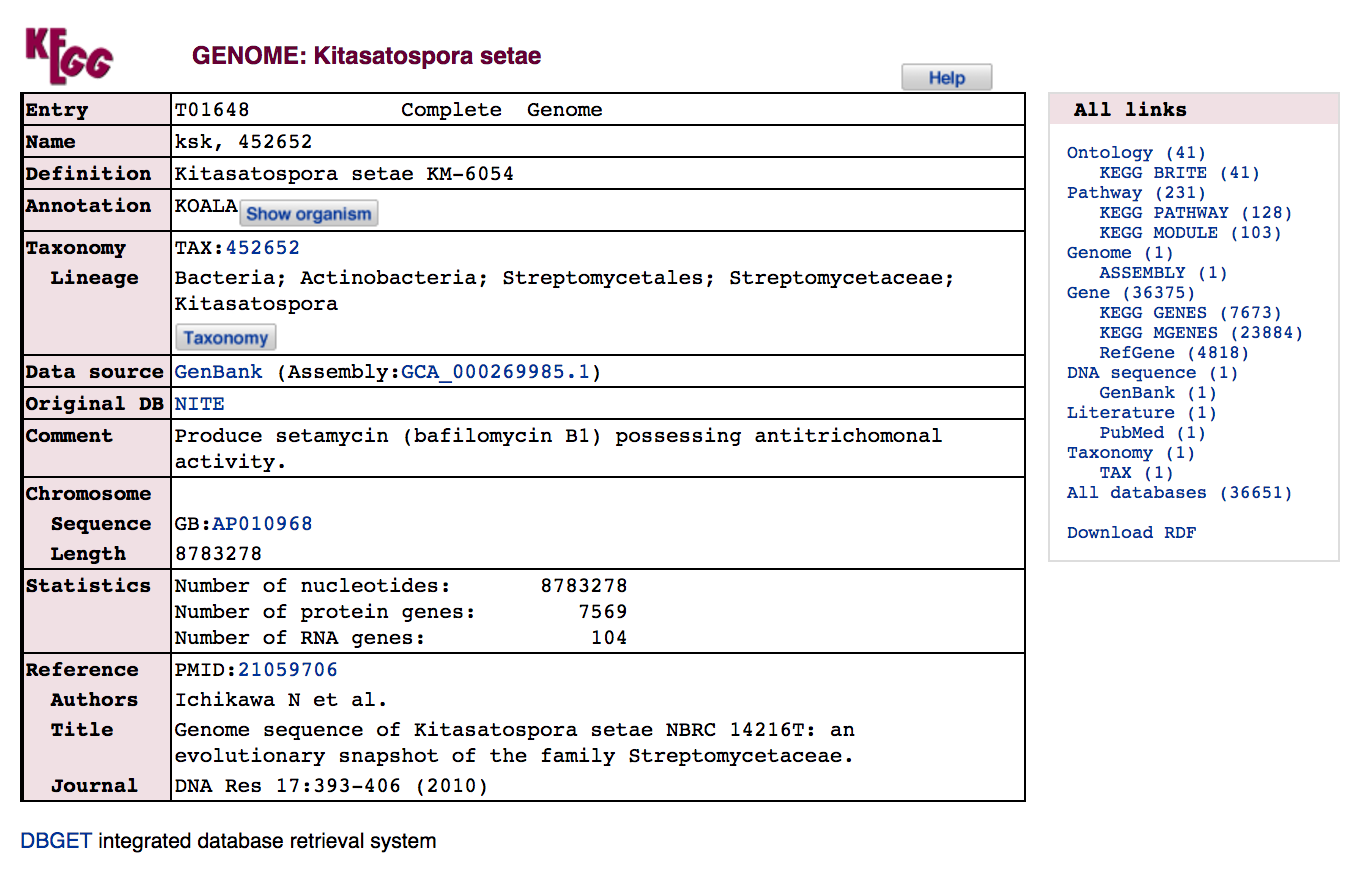
The KEGG entry for a genome typically links out to the GenBank record used for the KEGG entry, and also links internally to related genes and pathways in the other KEGG databases.
- Which `KEGG` databases are linked from the GENOME entry?
- How many Kitasatospora genes are annotated?
- How many `KEGG` GENES entries are linked from Kitasatospora genes?
- How many `KEGG` PATHWAY pathways include genes from Kitasatospora?
KEGG GENES¶
KEGGGENES: Molecular building blocks of life in the genomic space
When genes are imported into the KEGG system, several analyses are applied, including assignment to KO (KEGG Orthology) groups that attempt to cluster genes that have similar function, on the basis of sequence similarity. These KO groups have their own KEGG KO database:
KEGGKO: KO Database of Molecular FunctionsClick on the menu link to the GENES database.
Keeping the Kitasatospora genome with code ksk as our example from above, we will search for genes whose annotation includes the phrase xylulose:
- Set
Organismtokskand enterxylulosein the search field, then clickGo.
- How many gene entries are returned?
- How many different EC numbers are represented in the returned results?
We will select one of the returned genes to focus on: KSE_17560.
- Click on the link for
KSE_17560.
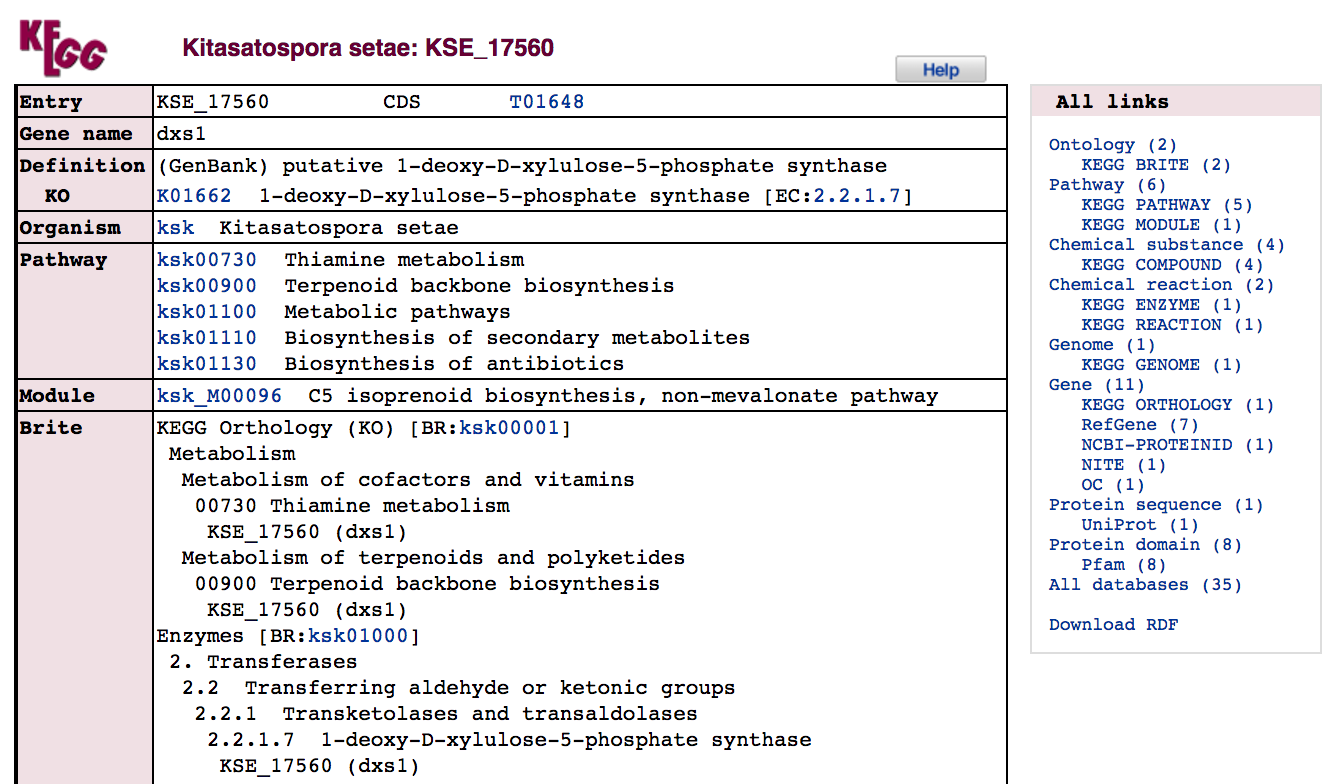
The gene record describes links to KEGG resources such as pathways, functional modules, and predicted orthologues, paralogues and gene clusters.
- How many `KEGG` PATHWAY pathways include this gene?
- How many `KEGG` MODULE modules include this gene?
The record also shows the amino acid and coding sequences (with optional upstream and downstream sequence) for the gene. Links to these sequences can be followed via the AA seq or NT seq buttons.
- Click on the AA seq button.
- How would you download this sequence?
KEGG PATHWAY¶
KEGGPATHWAY: Wiring diagrams of molecular interactions, reactions, and relations
The PATHWAY landing page gives a single search field, allowing a free-text search of the complete PATHWAY database. Note that the "Organism" selected by default is map, which is the generic reference map.
- Enter "terpenoid" in the search field and click "Go".
The search result lists pathways that match the free text search, and gives a short account of each.
- How many pathways are returned for this search?
- Click on
map00900.
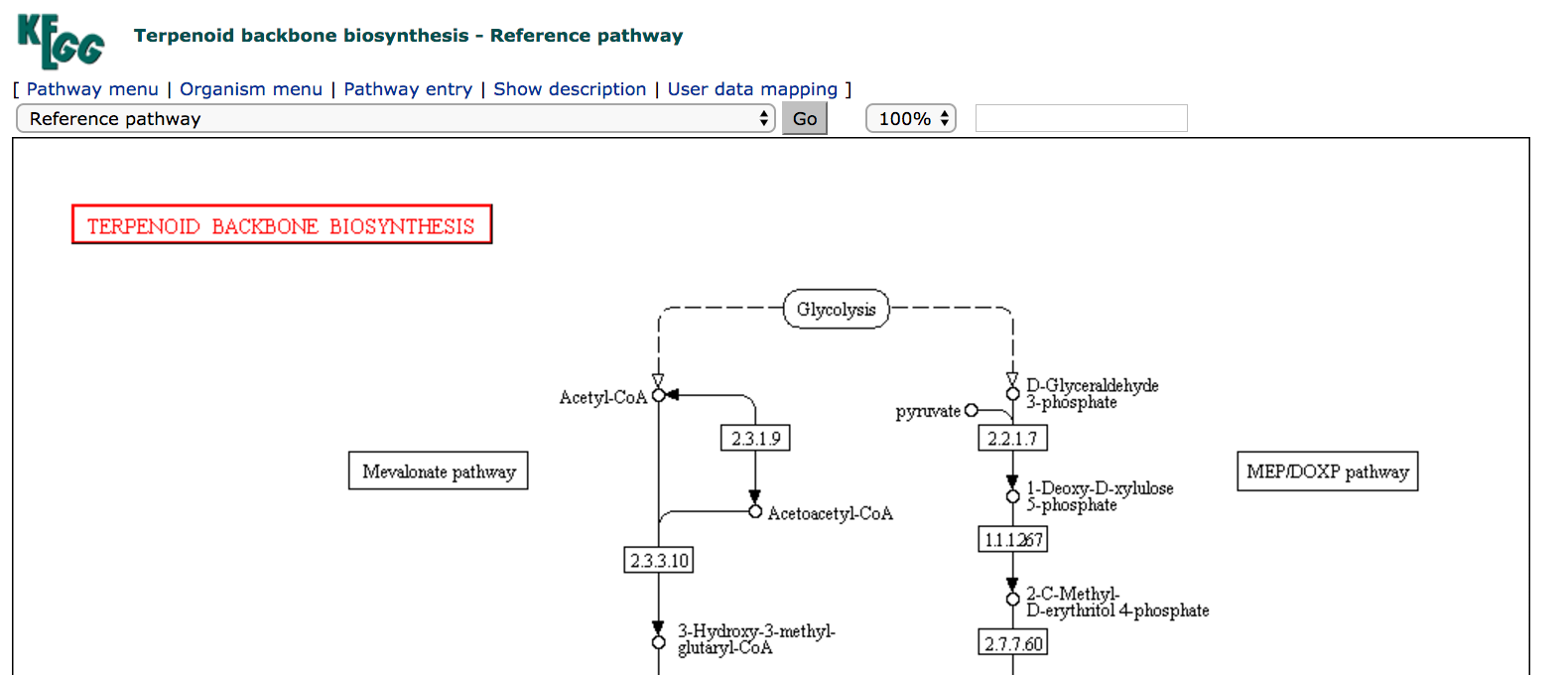
The map shown is a reference map, and is interactive:
- clicking on a rectangular node will take you to an entry in the
KEGGORTHOLOGY (KO) database - clicking on a circular node will take you to an entry in the
KEGGCOMPOUND database - clicking on a red node will take you to the connected entry in the
KEGGPATHWAY database
- Return to the
KEGGPATHWAY search page. - Enter
kskin the "Organism" field, and search again for "terpenoid".
- How many pathways are returned for this search?
Note that the search result pathways now have the prefix ksk. This is how KEGG allows us to index pathway maps directly: the three-letter code identifies the organism, and the five digit number identifies the pathway. Combining the two as <organism><pathway> means we can directly construct the pathway map ID for any combination of organism and pathway.
- The three-letter code for *Homo sapiens* is `hsa`. What would be the ID for the human terpenoid backbone biosynthesis pathway?
- Click on the
ksk00900map. This will open the terpenoid backbone biosynthesis map for Kitasatospora setae.
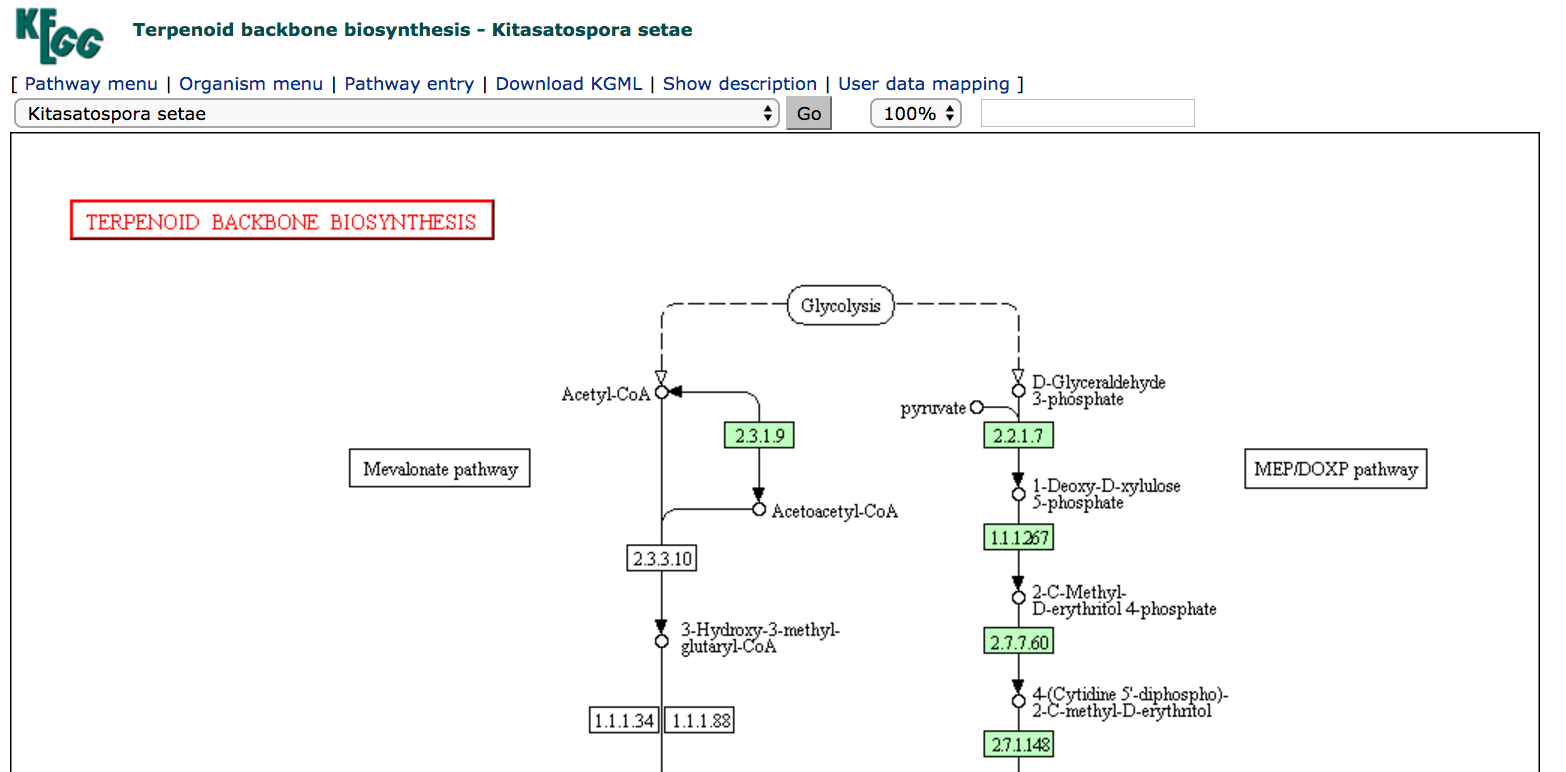
The ksk map is identical in structure to the map map, except that some of the rectangular boxes representing chemical interconversions have now been shaded green. This indicates that there is a gene annotated in the organism whose product is expected to be able to perform the biochemical interconversion indicated on the map.
- Click on the green box with
2.2.1.7.
- How many genes are linked from the box marked `2.2.1.7`?
"Special" maps¶
map01100: Metabolic pathways overviewmap01110: Biosynthesis of secondary metabolitesmap01120: Microbial metabolism in diverse environmentsmap01130: Biosynthesis of antibiotics
Following the links above, these are interactive. The individual pathways can be highlighted by selecting the checkboxes on the left hand side, and clicking on the circles will link out to compounds, while clicking on the graph edges will link out to reactions.
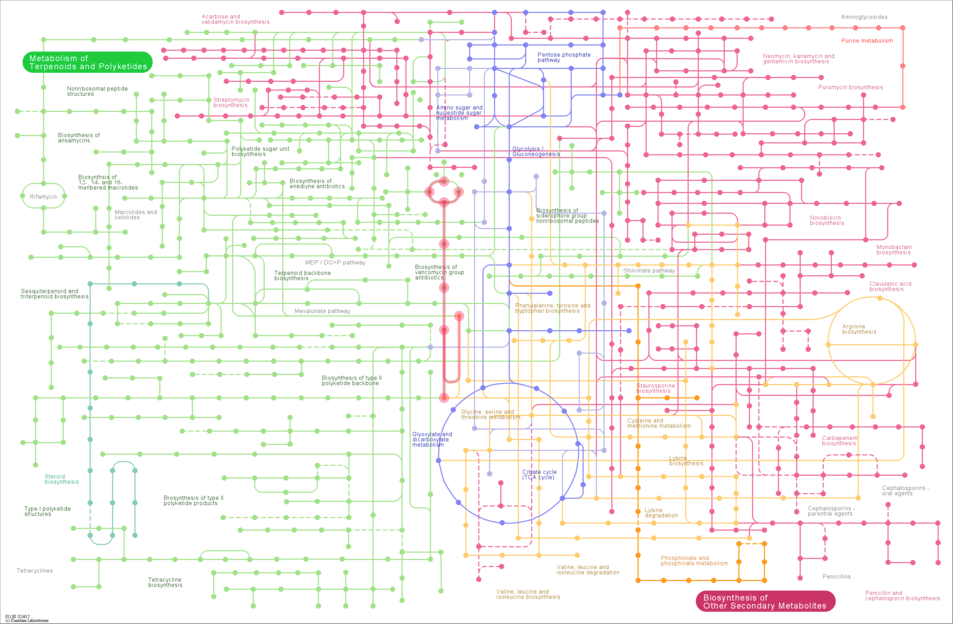
- Return to the main
KEGGPATHWAYS database page. - Click on the link to "Biosynthesis of antibiotics" in section 1.0 (Global and overview maps). This will bring up the interactive map.
- Select
M00774Erythromycin biosynthesis on the left-hand side. Note that the erythromycin pathway is highlighted. - Click on the "Reference pathway" drop-down menu. Scroll down to Kitasatospora setae, and select that organism. HINT: if you type "kitasat" with the drop-down selected, that will take you to the Kitasatospora option quickly.
- Click on "Go".
After a short delay, you should see that many of the edges and nodes in the image are greyed out. These pathways are not annotated in K. setae. The pathways that remain coloured are, however, annotated as present in that organism.
- Which antibiotic synthesis pathways/modules from `KEGG` are annotated as being present in *Kitasatospora setae*?
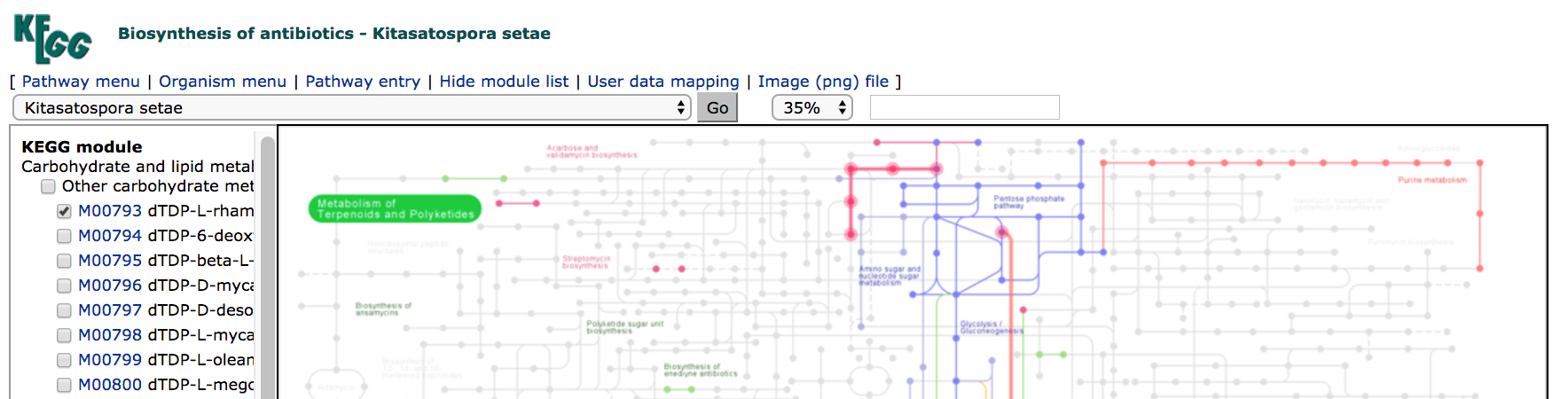
To download a copy of the image you have generated with your selections, click on the Image (png) file link at the top of the page.
Exercise 01 (15min)¶
The UniProt record Q05655 describes a human protein kinase. Using KEGG, can you discover:
- What is the record for this gene in the `KEGG` GENE database?
- Which pathways is gene product associated with?
- What diseases is this enzyme associated with?
- What are the substrates and products of this enzyme, and what is the `KEGG` REACTION database entry for the reaction?
- One of the pathways this enzyme is associated with is the chemokine signalling pathway. In the map for this pathway, what are the downstream pathways from this enzyme?
SOLUTION - EXERCISE 01¶
- hsa:5580
- 12 pathways
- Type II diabetes and insulin resistance
- ATP and protein; ADP and phosphoprotein; reaction
- Degranulation, Chemotaxis, NO induction, ROS production