10 - Ensembl¶
Introduction¶
Ensembl has a number of entrypoints or portals, divided broadly by type of organism:
Each of these resources is organised in a very similar way, and provides consistent browser, programmatic and pipeline-based routes to access their curated, integrated data. Not all features are available in each resource, however.
Resources¶
The EnsemblBacteria portal¶
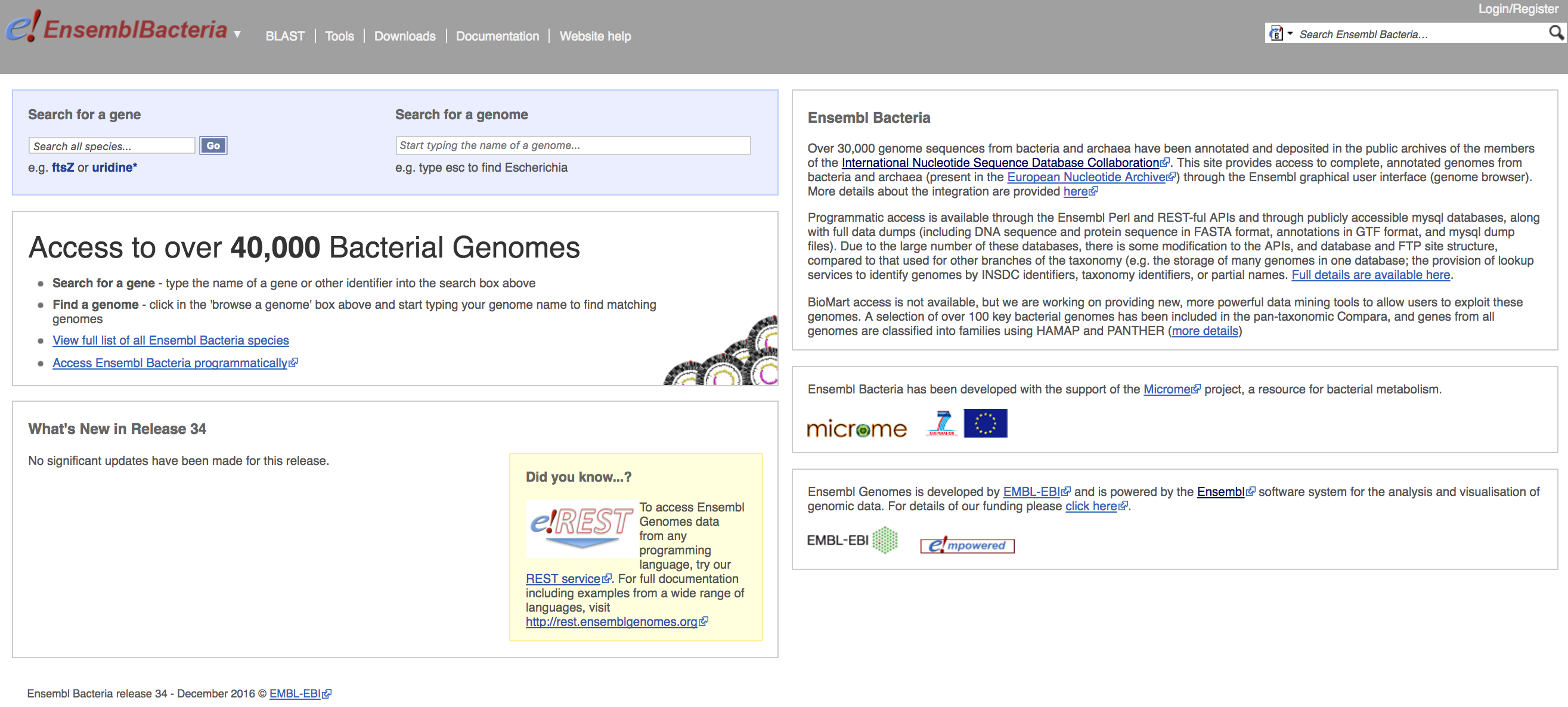
EnsemblBacteria provides access to over 40,000 bacterial genomes through a common genome browser interface. This data can also be accessed programmatically through a REST interface (see later lessons), and downloaded directly. Additionally, over 100 bacterial genomes are covered in the pan-taxonomic Compara tool. Bacterial genes from all the genomes are classified into families with HAMAP and PANTHER tools.
Given all this power behind the scenes, the landing page is deceptively plain:
Using Ensembl Genomes¶
As EnsemblBacteria provides over 40,000 genomes, it can be hard to navigate the full list, even though you can link to it from the landing page. It's much simpler to start typing the name of the species you want in the Search for a genome box, as for the term Pectobacterium below:
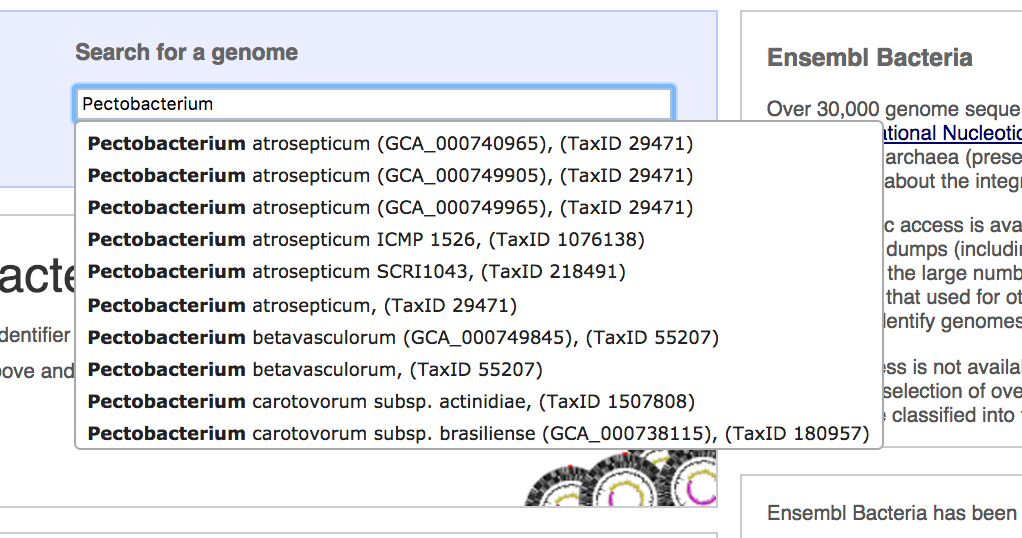
- Start typing Pectobacterium in the search field
This produces a drop-down list, which we can scroll through until we find our organism of interest. In this case, we're going for Pectobacterium atrosepticum SCRI 1043
- Click on the entry for Pectobacterium atrosepticum SCRI 1043
This brings up the genome's homepage, which offers several useful links for information and statistics, comparative genomics, downloading data and, at the top, another search bar so you can search for features of interest:
Downloading data¶
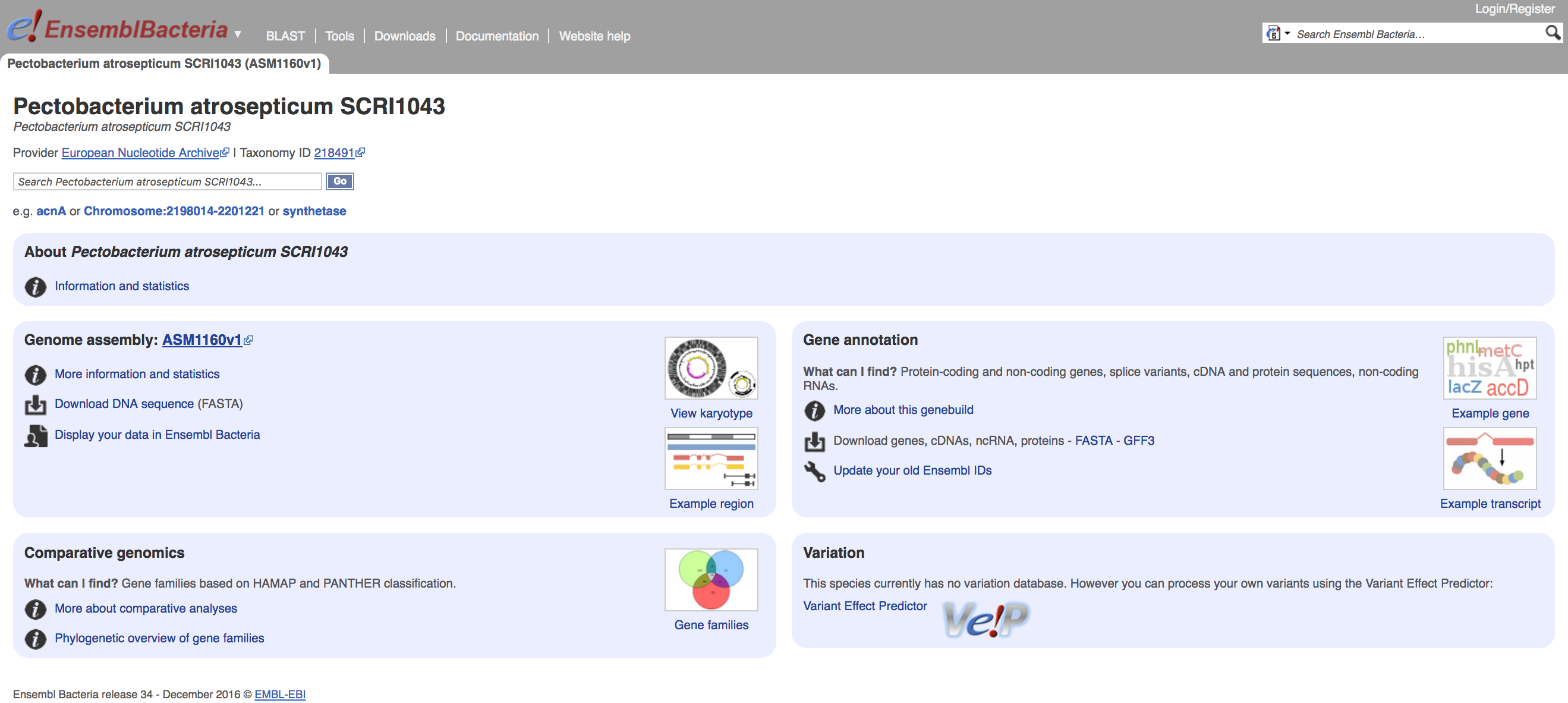
On the right hand side of the page, there are links to download gene and protein sequences in FASTA or GFF3 (usable with Artemis or Tablet) format. These also take you to an FTP site from where you can download the corresponding data.
The Genome Browser¶
- Click on the region of interest link to
Chromosome:2198014-2201221
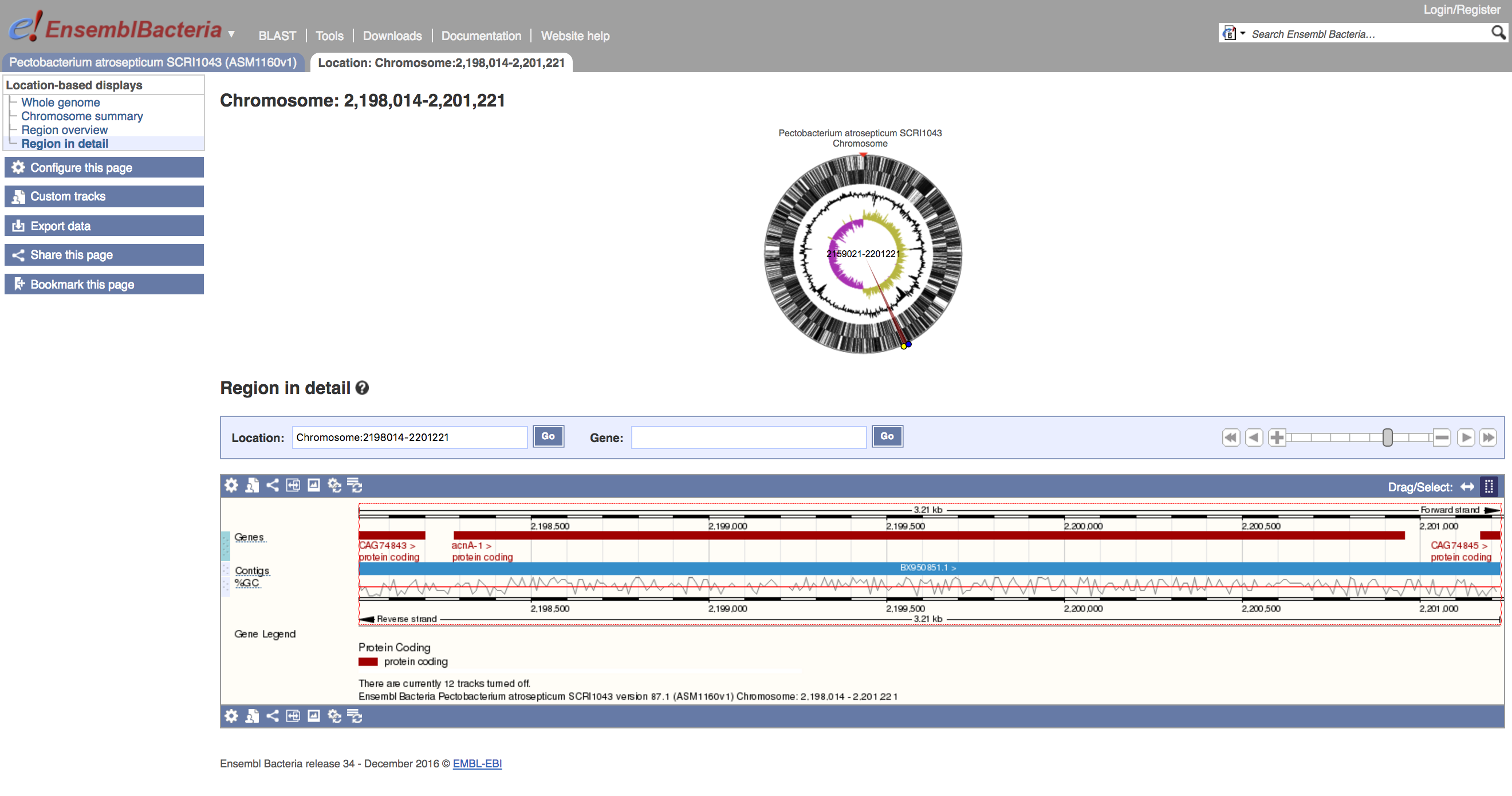
At the top of this page, there's a circular overview of the entire chromosome, with features and GC content/skew indicated, and the region of interest marked with a red wedge. Below this there's a linear view of the region in detail.
On the circular view, there are yellow and blue handles. You can click and drag these to modify the region of interest, and if you click on this new region, the linear view beneath the circle will update (if the region is not too large to view).
- Use the handles to zoom in on a region approximately 1,644434-1,688,332
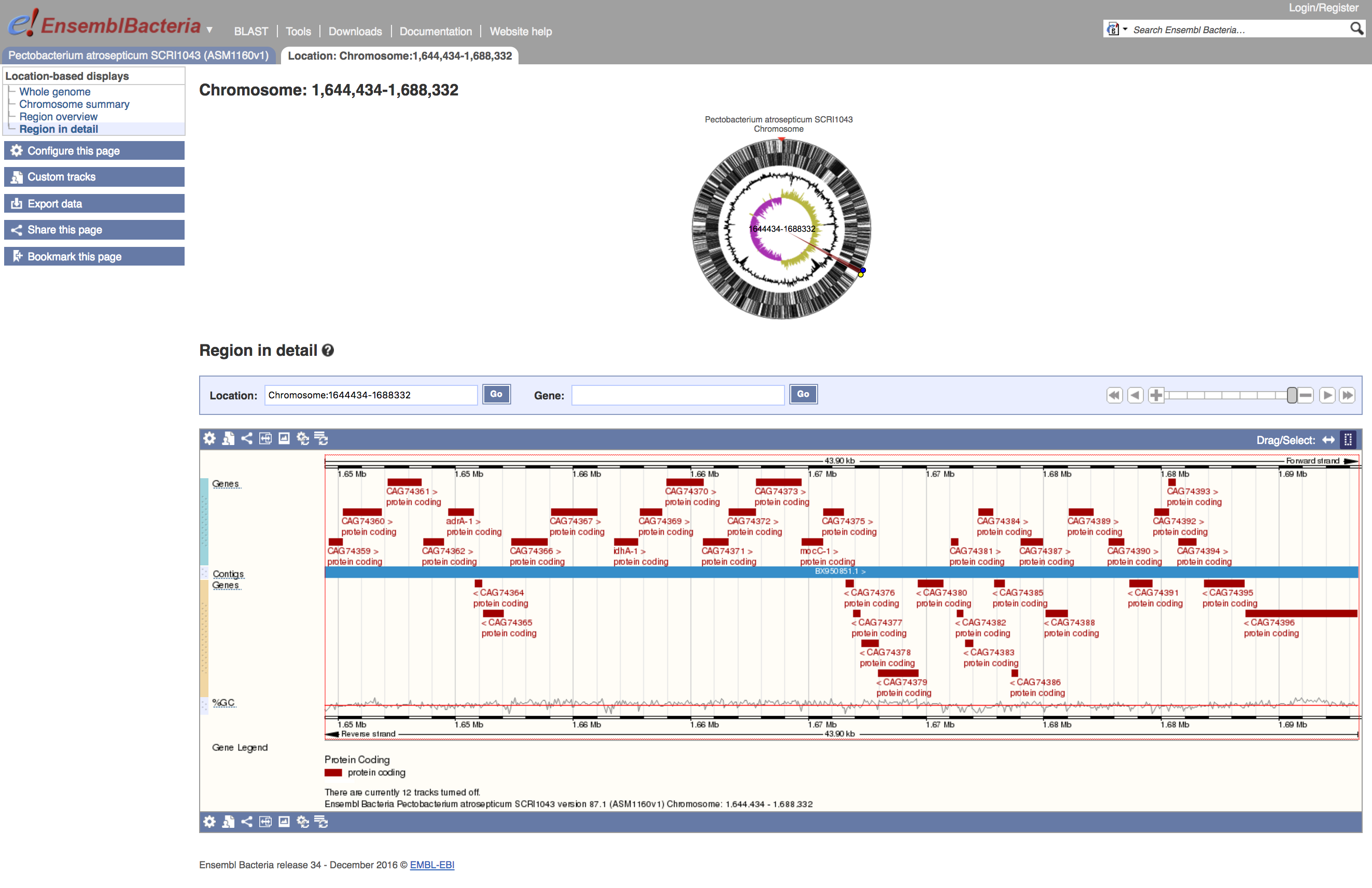
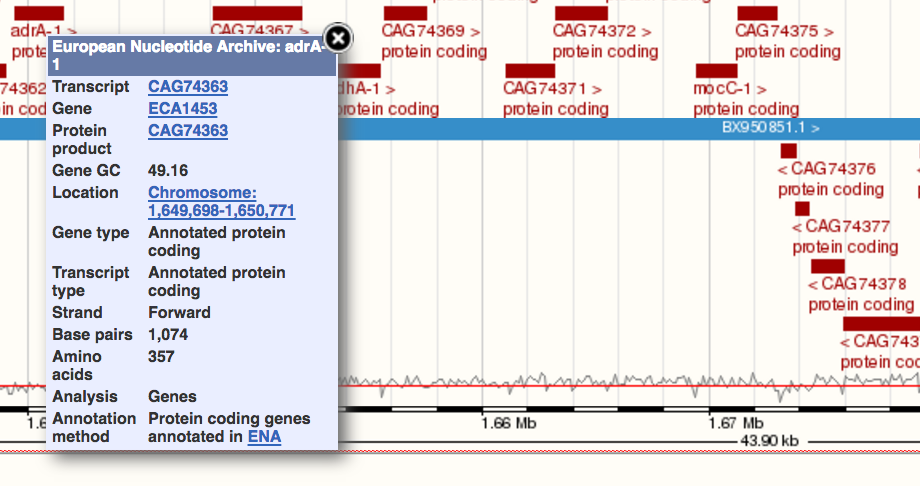
The lower region shows a view of the features annotated on the genome, coloured by type. In the example above, all the features are protein-coding genes. If you click on one of these, a small popup window appears, giving a little more information.
- Click on one of the features in the lower view
Gene view¶
In the popup you saw above, you will have noticed several distinct identifiers - for the transcript, the gene, and the protein product. These are live links, and will take you to a new page centring the browser on that feature, with an overview window that shows the gene-based display. For instance, clicking on the ECA1453 gene link above brings up the gene view:
- Click on the ECA1453 link
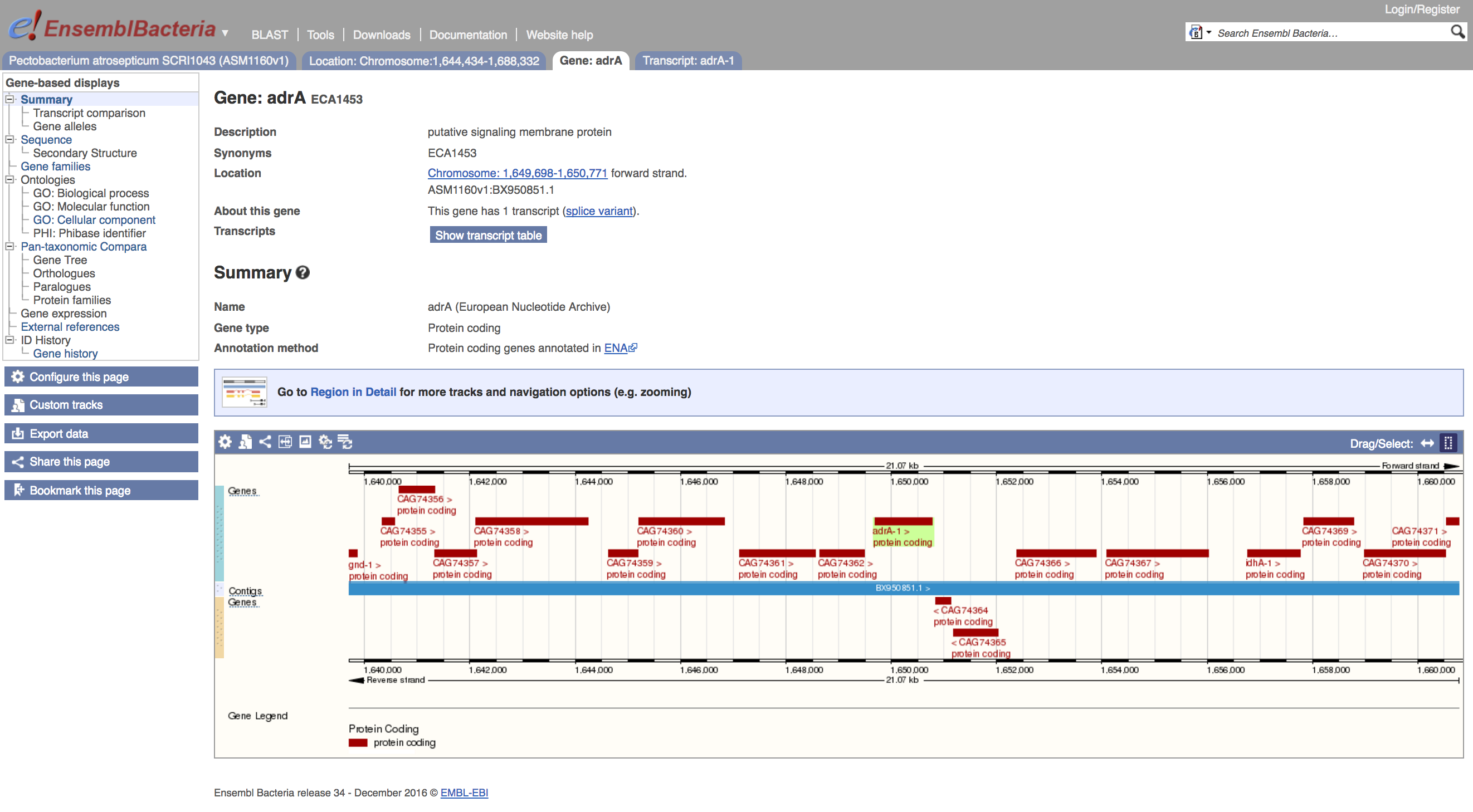
You will see that the content of the left-hand menu has changed, giving links out to sequence annotations such as GO terms, and external references.
To obtain an account of the links to other databases for this feature, you can use the External references link in the left-hand menu. This will bring up an account of other entries for this sequence.
- Click on the External references menu item
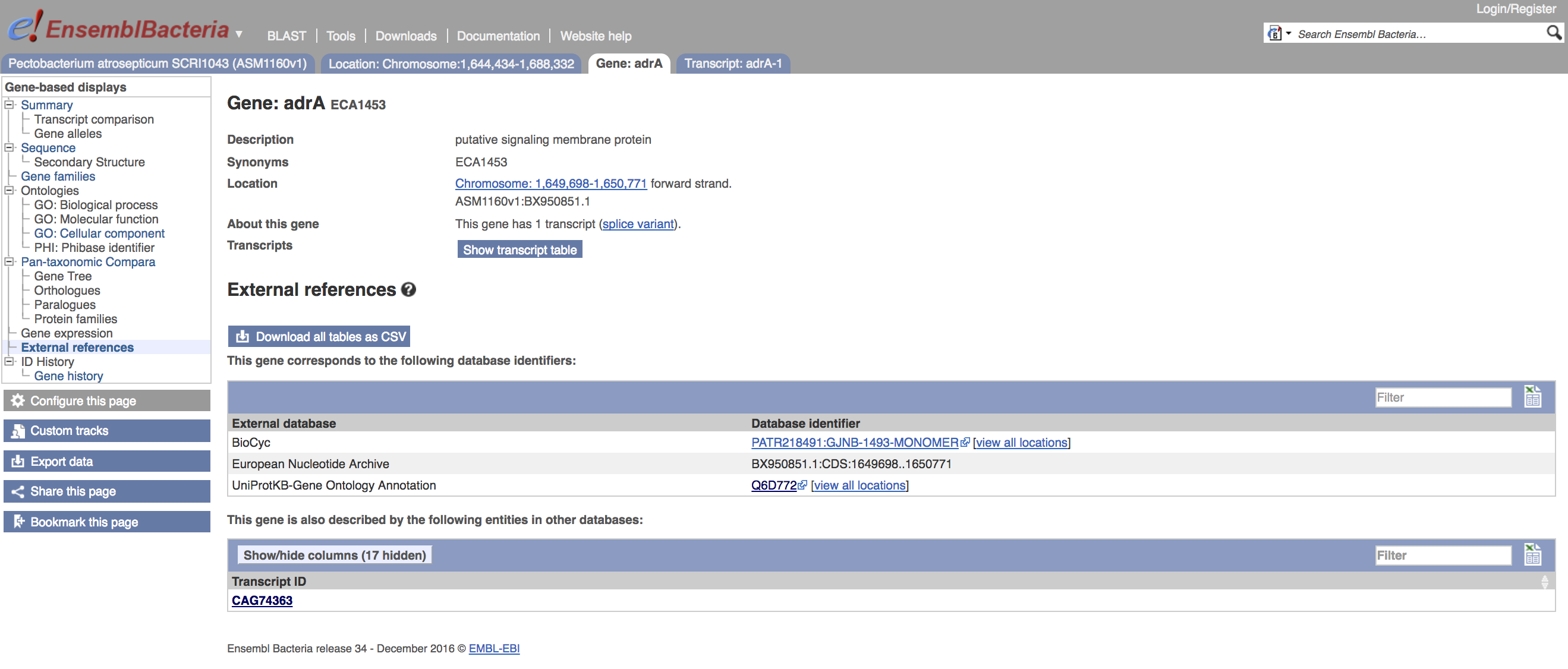
- Can you find the corresponding entry for this feature in NCBI/GenBank, using your browser?
You can search directly for a gene of interest in the the gene search box, and we will do this for the virulence-related protein VgrG, below.
- Click on the tab for the whole genome home page
- Enter vgrG into the search field and click
Goor hitReturn
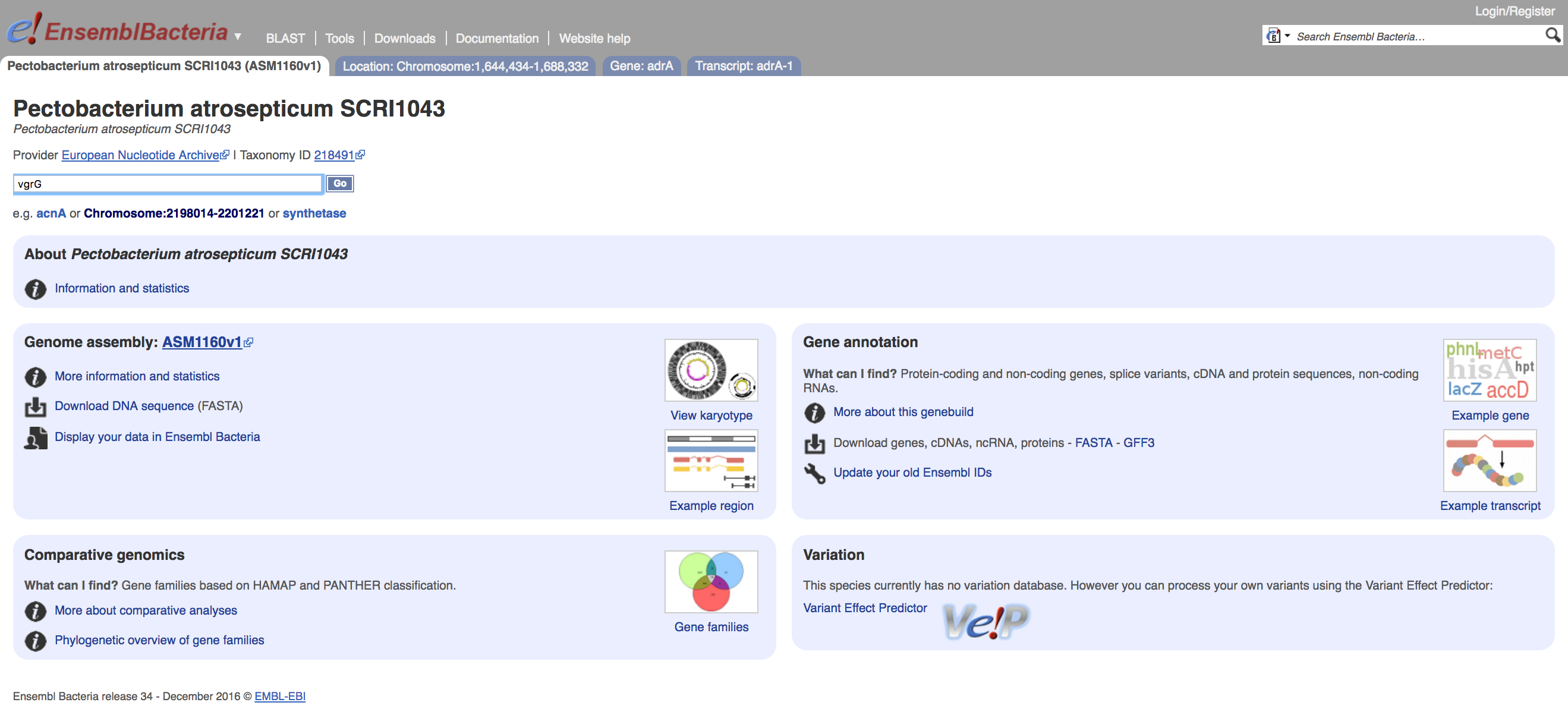
- How many genes does the search return?
- Are all of these results likely to be active coding genes?
- How many genes in the GenBank record for this genome are annotated as `VgrG` HINT: the genome record can be found here.
- Go to the gene page for the first hit, and click on
Sequencein the left-hand menu
This will present a marked-up view of the genomic sequence for, and flanking, that vgrG search result. There are buttons that will allow you to download the sequence in a range of formats, or submit the sequence as a BLAST search, and the exons are highlighted, with the current gene of interest shown in red.

The FASTA header shows the location of the region covered by the sequence.
Transcript view¶
- Click on the transcript tab
This will bring you to the transcript summary view:
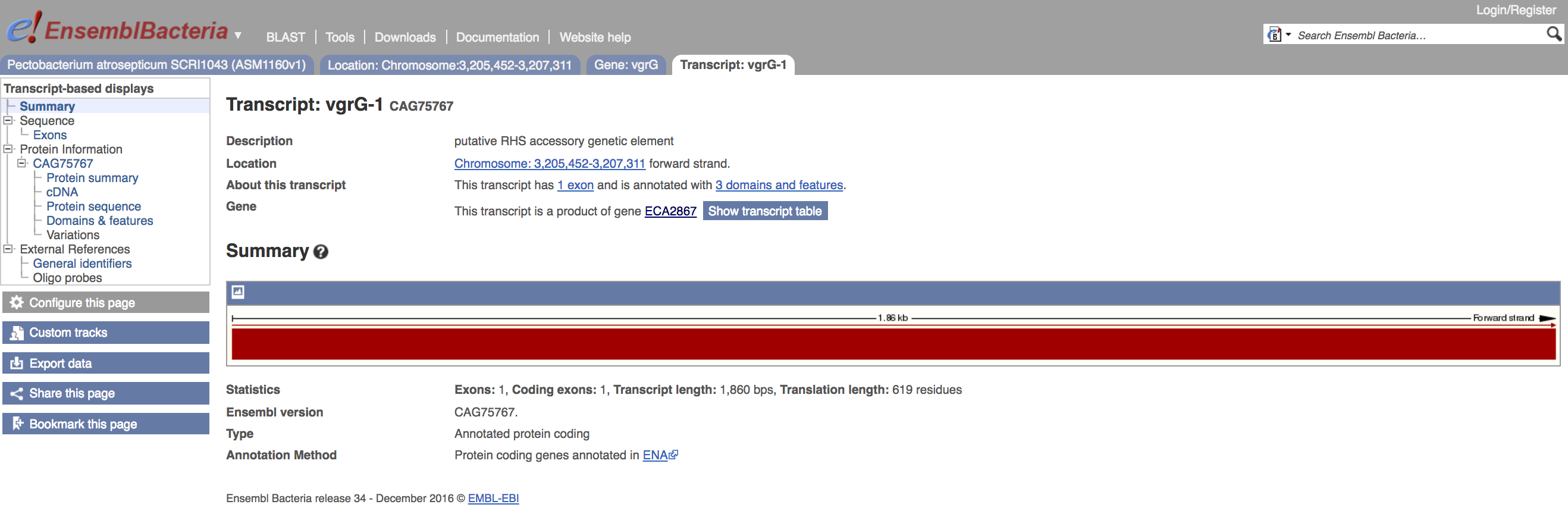
- Click on the
cDNAmenu item on the left-hand side
This will show the transcript coding sequence.
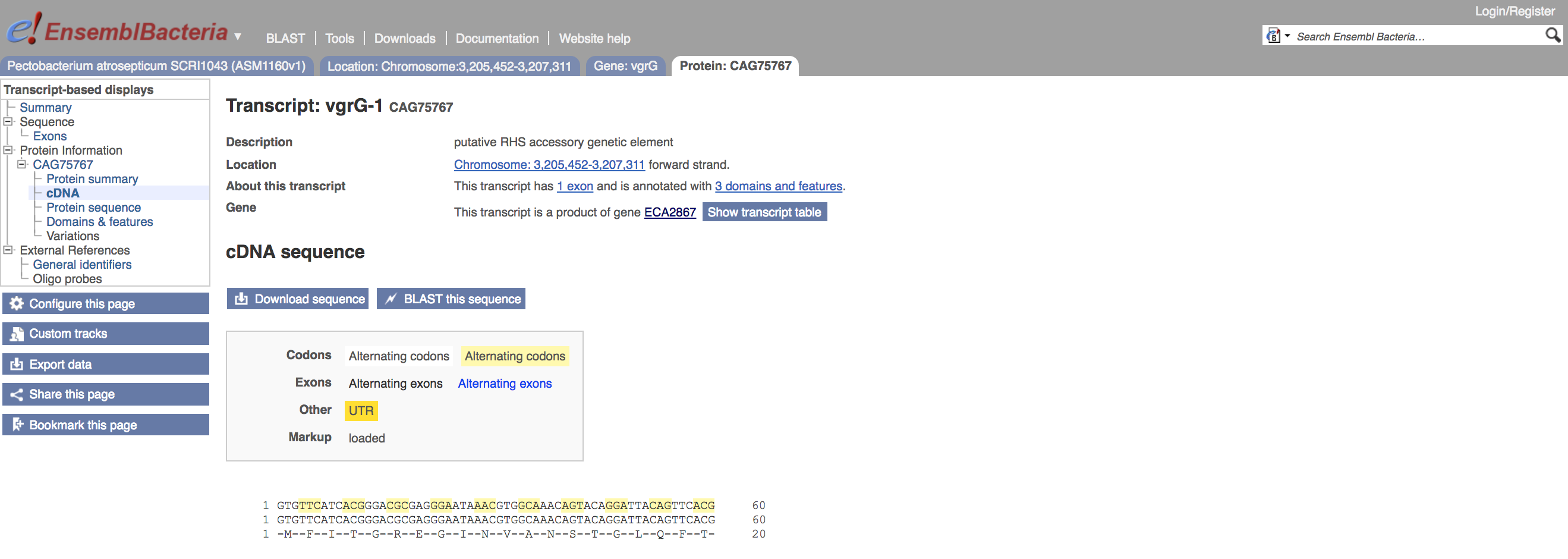
- Click on the
General identifiersmenu item
This shows the external database links, to find more information about this sequence. In particular, the link out to UniProtKB indicates the percentage sequence identity to one of UniProt's protein sequence entries. Here it is, as would be expected, a 100% identity match.
Comparative Genomics in Ensembl¶
Go to the page for E. coli K-12 MG1655
- Search for the gene pfkA.
- Click on the link for
b3916to get the gene view
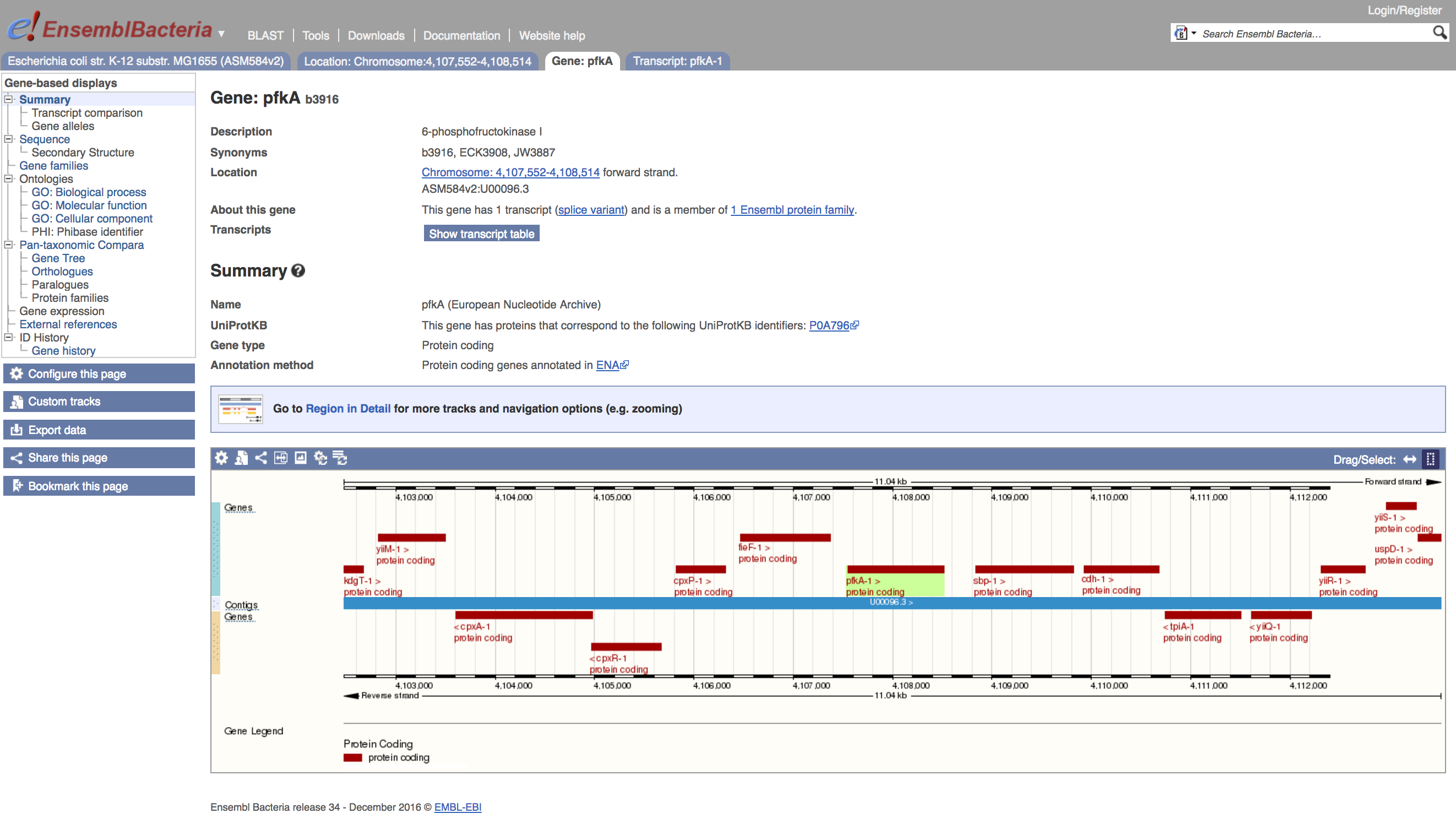
Note that the menu on the left now has the Pan-taxonomic Compara, Gene Tree and Orthologues links available. These are the entrypoints to Ensembl's comparative genomics tools.
- Click on the
Gene Treemenu option
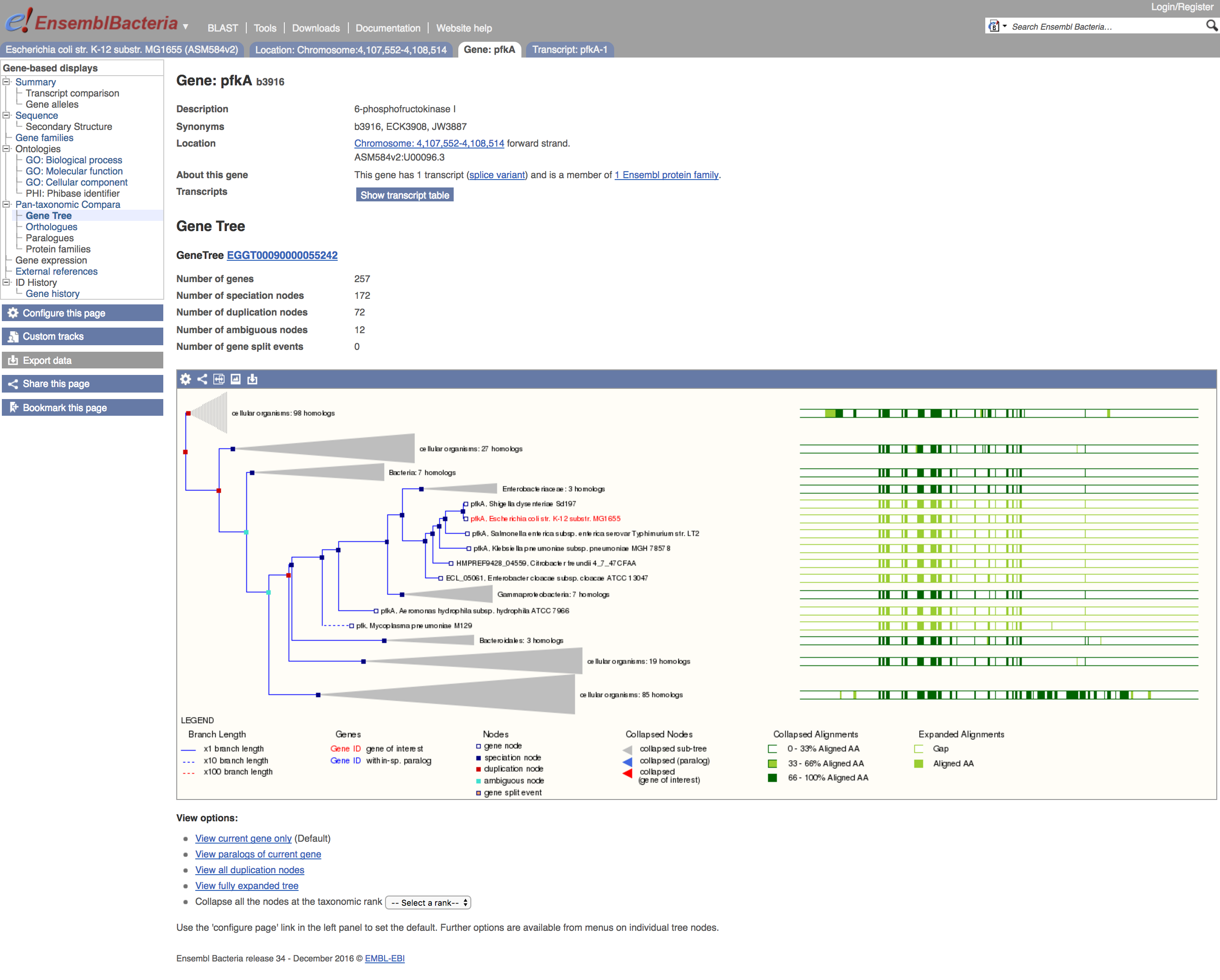
The browser now shows a tree locating this E. coli pfkA in the context of orthologues from other organisms. The E. coli gene of interest is highlighted in red, and a set of figurative protein alignments is shown to the right of the tree - these alignments indicate regions of the sequence and their extent of sequence identity. On the tree itself, several branches are 'collapsed' into shaded triangles, and more information about these collapsed sequence sets can be obtained by clicking on the grey triangle, to bring up a pop-up window.
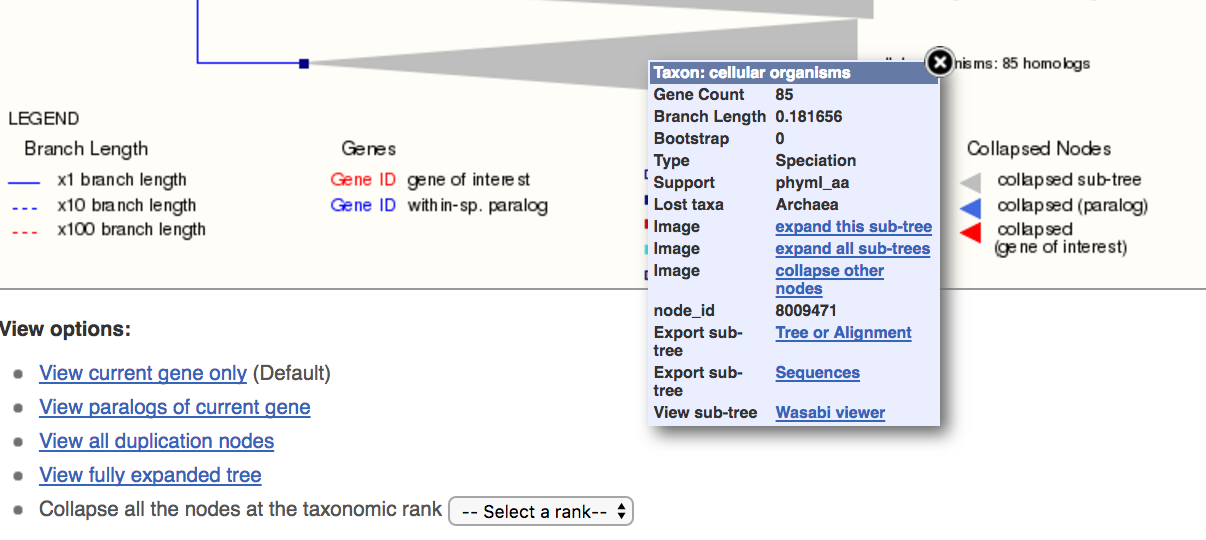
The pop-up has a link (expand this sub-tree) that allows you to see the members of the collapsed branches in full.
- Click on the
Orthologueslink in the left-hand menu
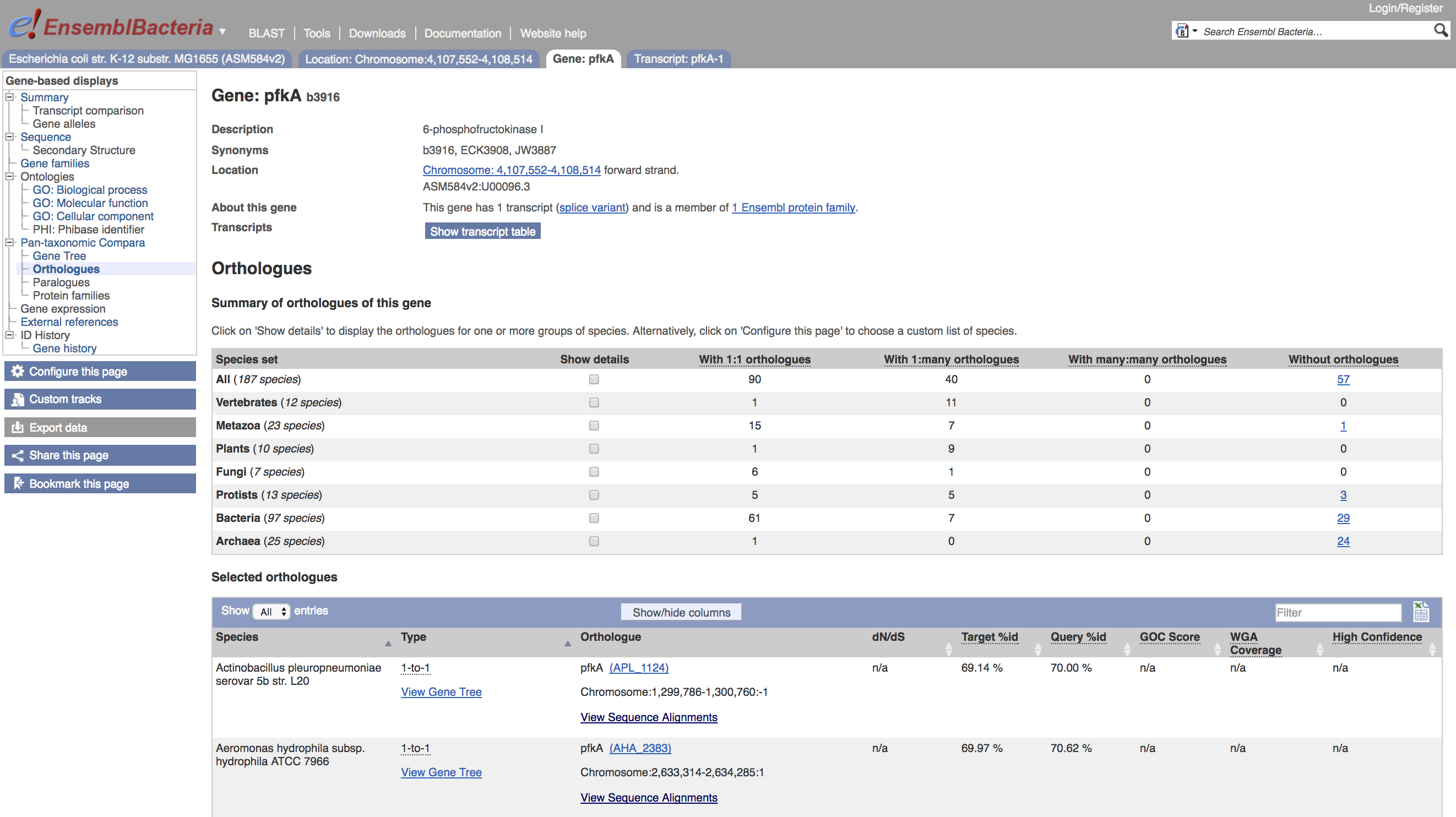
This will present the set of orthologues to pfkA in the Pan-taxonomic Compara as a table:
- How
Ensemblcalculates and describes orthologues: Compara homology methods
The table is interactive and will allow you to live-filter on organism type by selecting Show details for any of the offered groups. Also, each individual row in the table presents links to view the corresponding gene page, and the sequence alignment.
- Click on
Show detailsto restrict the table to bacterial orthologues - Enter
Mycointo the table filter to restrict the table to two rows - Click on the link to view sequence alignments in the first row
Clicking on the sequence alignment link will bring up a pop-up menu asking if you want the protein or cDNA alignment.
- Click on
View Protein Alignment
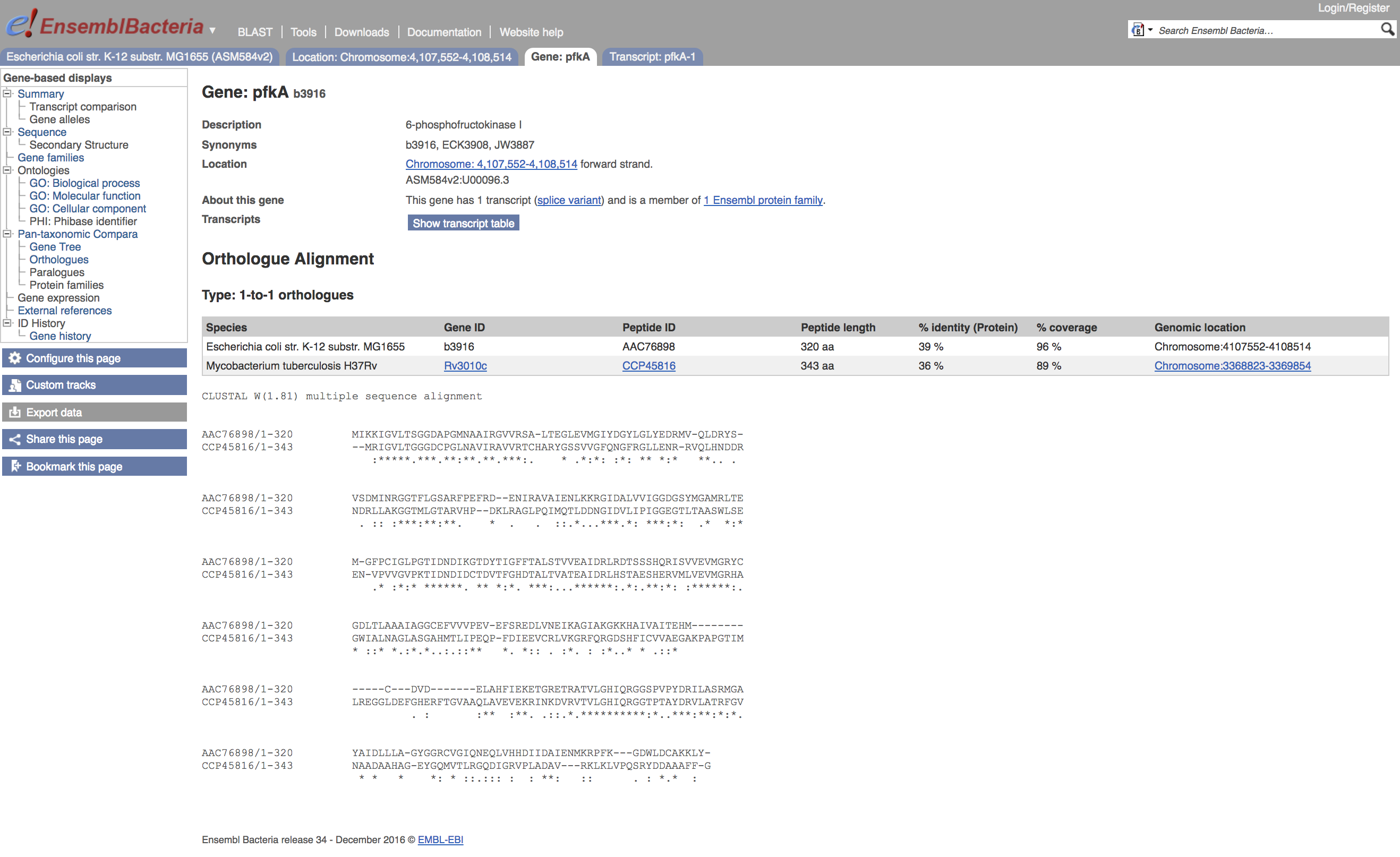
This gives the pairwise alignment used in constructing the orthologue set for your query sequence, in ClustalW format. There is information given on sequence percentage identity and coverage for this alignment.
Exercise 01 (10min)¶
Using the EnsemblBacteria tools, starting from EnsemblBacteria, can you:
- Go to the home page for the *Kitasatospora setae* KM-6054 genome
- Find how many coding genes are annotated in this genome?
- Download the FASTA file describing all predicted protein gene products?
- Find how many putative penicillin-binding proteins are annotated in the genome?
- Examine the feature `KSE_59840` in the genome browser, and find the GO term for its molecular function? What is the evidence for this functional annotation?
- What is the `UniProt` accession for this protein?