01 - Searching in the RSCB for PM lipase structures¶
Table of Contents¶
Introduction¶
We will use the RCSB to search for the structures of PMLs and explore their 3D structures. Its is possible to search the RCSB in a number of ways, including by PBDcode ID and sequence similarity.
From the previous session on PML sequences it is possible to find that PDB code ID for the wildtype (WT) PM lipase is 4GW3 (there were cross references to the RCSB from the Uniprot entry: B4EVM3_PROMH)
Searching the RCSB¶
Use the search box at the top of the RCSB homepage to search for entry 4GW3.
This shows the "Structure Summary" page for 4GW3
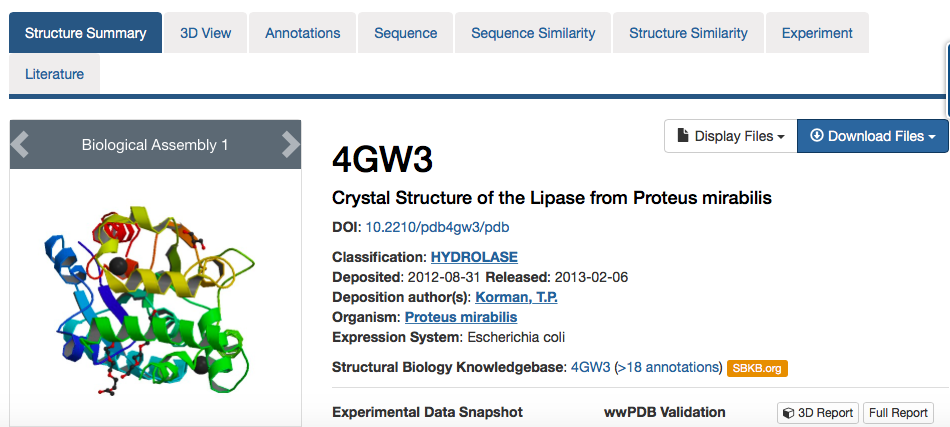
Note the different tabs at the top of the entry which give access to difference types of information about the 4GW3 structure.
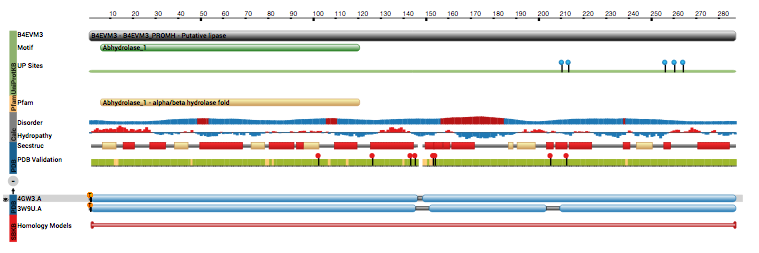
Using the expanded Macromolecules section of the Structure Summary page,

we see that this structure has one Pfam sequence domain and we will come back to view this domain on the structure later.
Exercise 01: [5 mins]¶
Browse the RCSB 4GW3 entry and answer the following questions
- What method has been used to solve the structure of 4GW3 and is the "quality" of the structure suitable for analysis of its ligand binding sites?
- How many amino residues does this PML structure have?
- What ligands does this PML structure have bound?
- What is the protein stoichiometry of 4GW3?
- Which Pfam sequence domain is present in this protein and what residues does it span?
- What other proteins in the PDB share 95% sequence similarity with 4GW3 and do you think that these proteins have the same structure and function?