02 - Visualization of PML structures using a web-based tool (NGL)¶
Table of Contents¶
Introduction¶
In the first practical (4-01) the sequence similarity tab in RCSB showed the sequence similarity clusters of 4GW3. These clusters showed 4 structures that have 95% sequence identity.
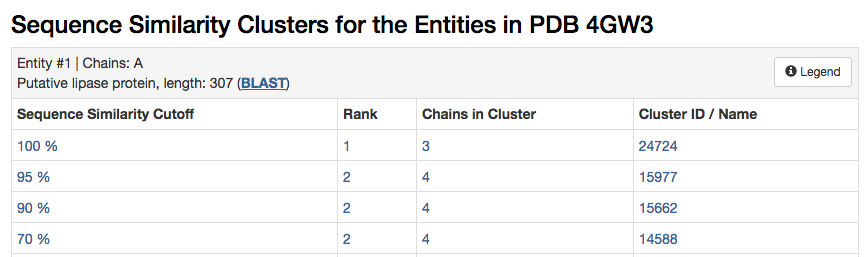
| RCSB Code | Structure Description |
|---|---|
| 4GW3 | PML- wild type (WT) |
| 4GXN | PML -WT Diethylphosphonate Inhibited |
| 4HS9 | PML Methonal tolerant mutant |
| 3W9u | PM Lipk107 |
We will concentrate on the first 3 structures, as the 4th is poorly annotated and does not have a publication associated with the RCSB entry. Hence it is more uncertain what this structure actually is.
Using NGL¶
The RCSB supports 2 web-based applications (NGL and JSmol) to allow visualization of protein structures directly from the website.
Click on the 3D View tab at the top of the 4GW3 entry to view the structure. Ensure the NGL application is being used to display the structure (use the "Select a different Viewer" box to select NGL(WebGL).
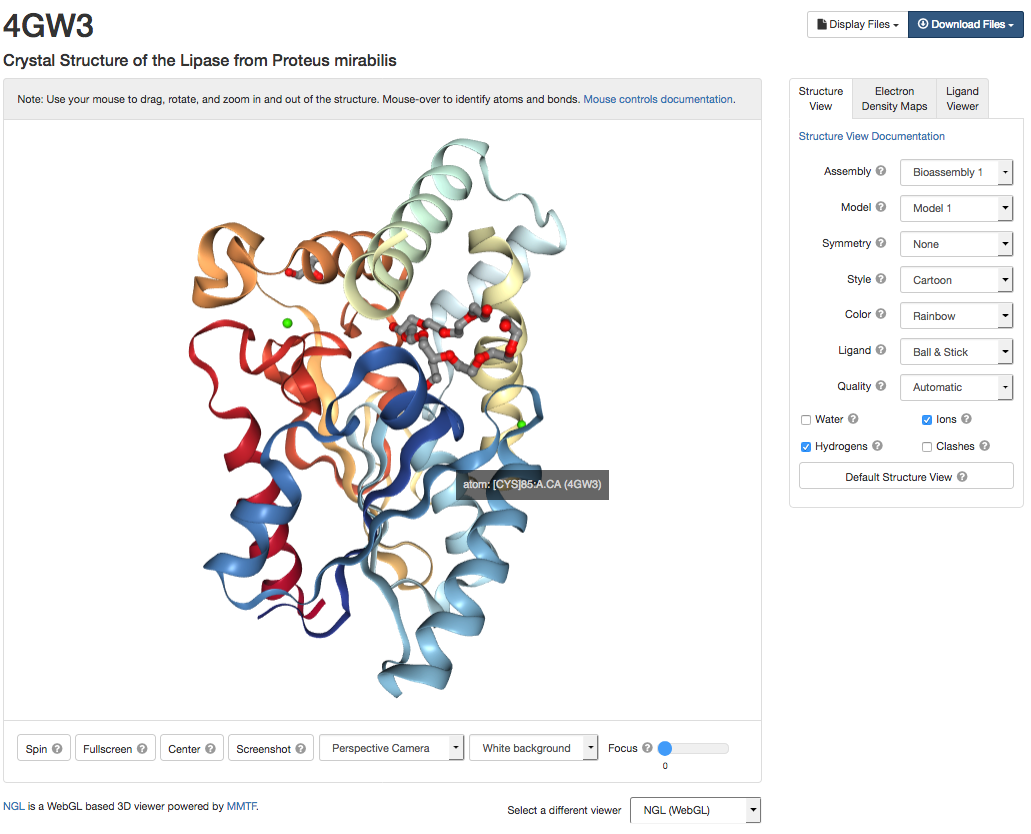
You can use the "Structure View" tab on the right hand side to alter how the protein and its ligands are drawn. The colour scheme options can be used to display the surface of the protein in different ways E.g. colour by hydrophobicity.
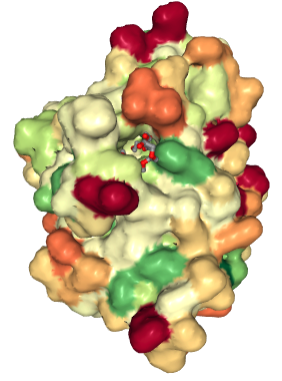
If there are ligands or ions in the structure you can use the "Ligand Viewer" tab on the right hand side to display the ligands in their binding pockets with their interactions. e.g Binding pocket for Pentaethylene Glycol (PEG)(1PE).
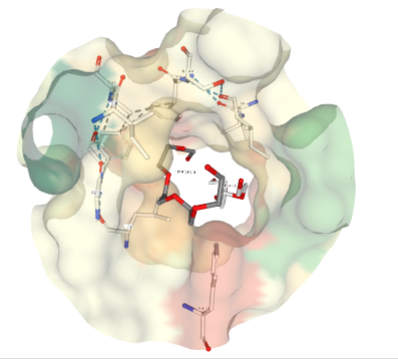
Exercise 01: [10 mins]:¶
Use the ''Display and Viewer Options'' on the right hand side to:
- colour 4GW3 by secondary structure
- show the position of calcium ions in 4GW3
- view the surface of the protein and colour by hydrophocity
- view the surface of the protein and colour by b-factor
- What fold does 4GW3 have? Search the [CATH database](http://www.cathdb.info/) to help you answer this.
- What amino acids residues are involved in binding Calcium ion CA 401?
- What does the hydrophobicty colour scheme show you about the binding pocket where the ligands PEG occurs?
- Each atom of protein crystal structure can have a B-factor (also known as the temperature or Debye-Waller factor ) associated with it. This essentially describes the degree to which the electron density is spread out. When you colour the protein surface by this factor what do you think it shows you?