

UniProt (browser)¶UniProt is a comprehensive protein sequence and annotation resource, and is a consortium between the European Bioinformatics Institute (EMBL-EBI), the Swiss Institute of Bioinformatics (SIB), and the Protein Information Resource (PIR). UniProt provides unifies several legacy databases, uncluding Swiss-Prot, TrEMBL, iProClass and the PIR-PSD.
UniProt provides three key databases:
UniProtKB is likely to be the database you use most frequently to find information on gene product/protein molecular function. It is the central hub of functional information on proteins, and collates functional annotations from many other databases, ontologies, and references. It keeps records of how annotations are derived (e.g. experimentally or computationally), with evidence codes, and is divided into two sections: one contains manually-annotated records (UniProtKB/Swiss-Prot), and a second contains computational annotations that are waiting for manual curation (UniProtKB/TrEMBL).
UniParc is a comprehensive, non-redundant database that contains most of the publicly-available protein sequences from a range of sources.
UniRef provides pre-clustered sets of sequences from UniProtKB and UniParc. A number of clusterings at different stringencies are provided.
These databases can be queried in a number of ways, including:
UniProt website http://www.uniprot.org/ in your web browserUniProt website, using a programming languageUniProt website¶UniProt webpage.
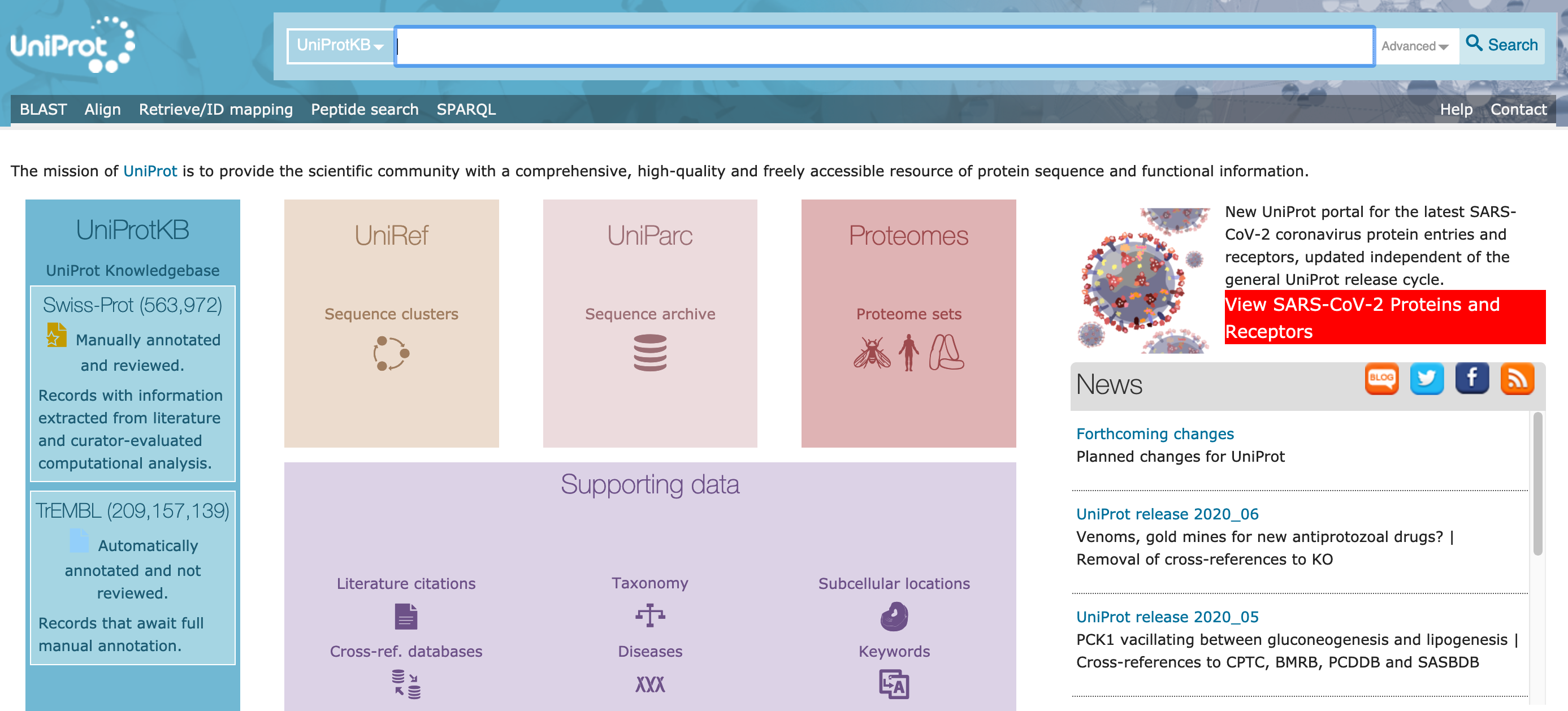
The landing page offers options for each of the three main databases: UniProtKB, UniRef, and UniParc. It also offers sets of complete proteomes for a range of organisms, and databases of proteins organised by supporting data, such as literature, taxonomic classification, and subcellular location.
UniProtKB¶UniProtKB link. This will take you to the UniProtKB front page, with a summary of entries, and a number of links.
Search button.

"kitasatospora" as an organism or by taxonomy. Click on the `organism` filter.
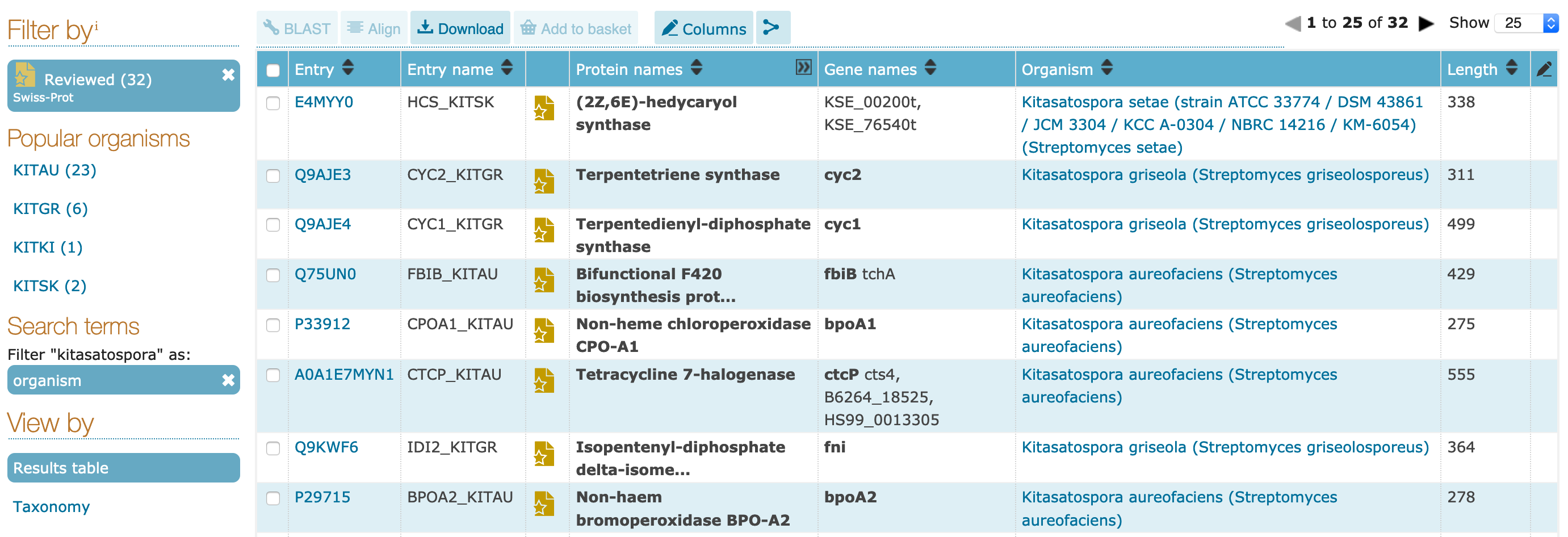
Q9AJE3.

History. Click on this button. A small window will open, with a link to Previous versions. Click on this link.

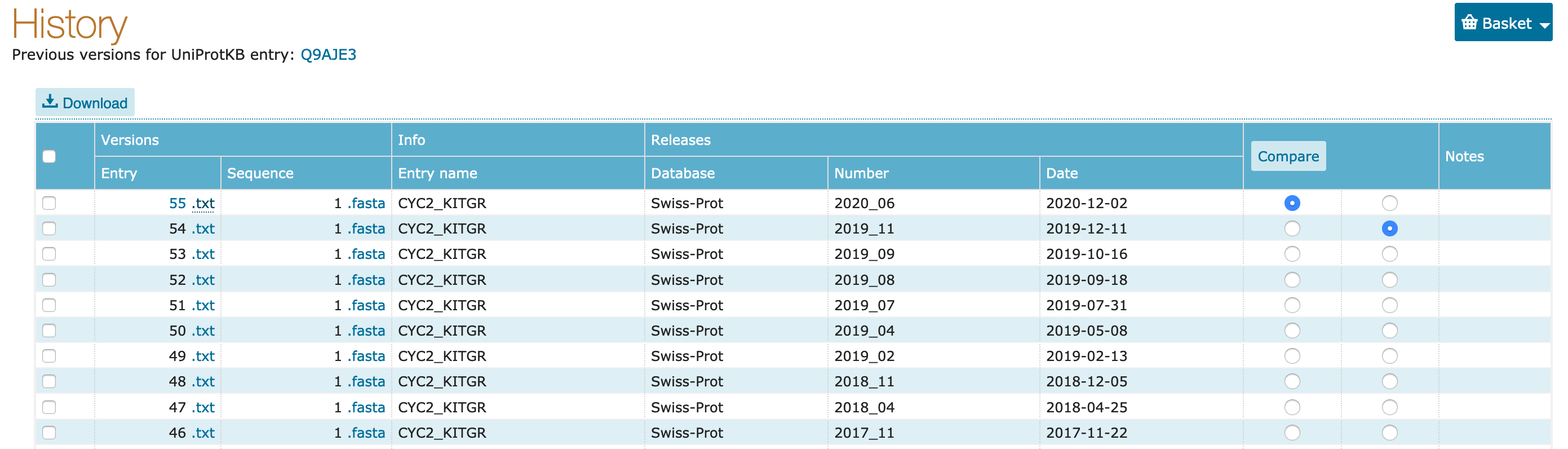
Compare button.
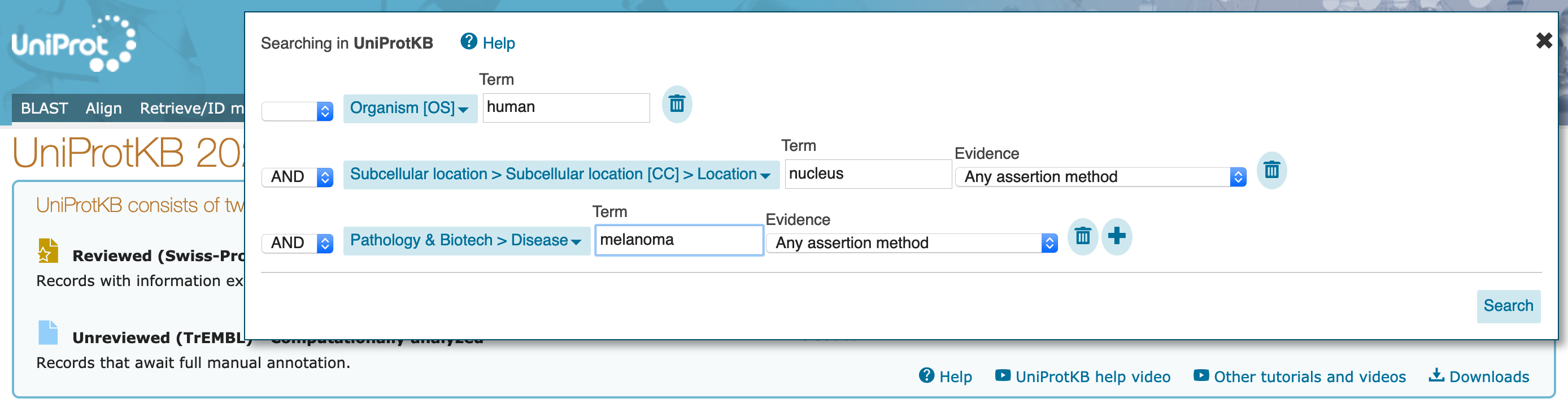
In this section, you'll use the advanced searches to identify candidate human proteins that are found in the nucleus, and have been associated with some disease activity or function.
UniProt logo to return to the landing page
"Advanced" drop-down to get the advanced searching interface
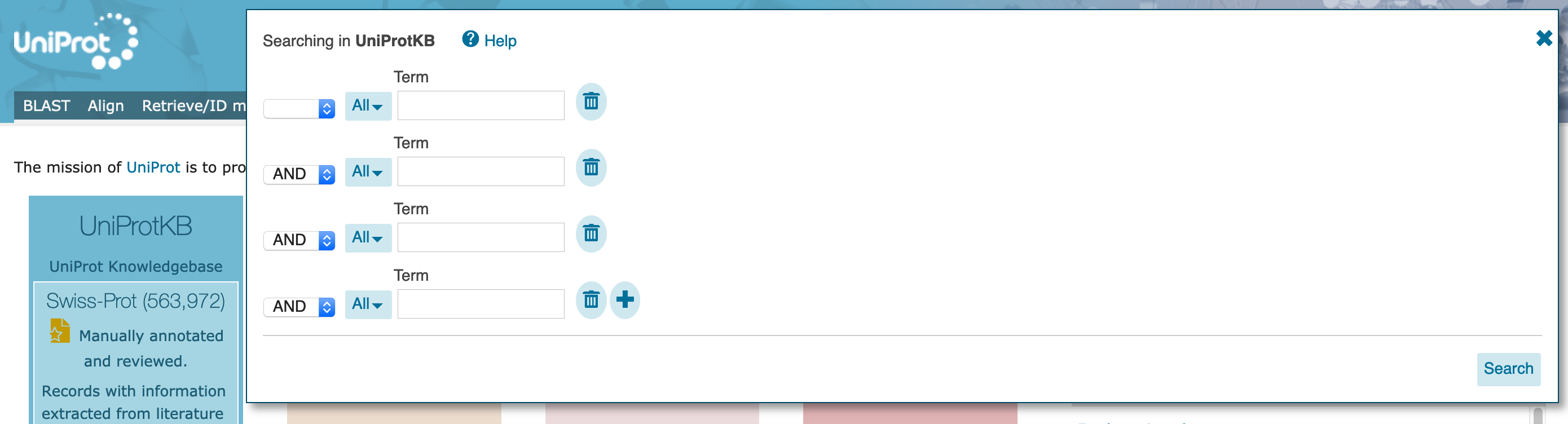
Organism [OS] with search term "Human". The dropdown will offer you several options as you type, but do not select them (you could have entered the organism "Homo sapiens" here, also).
AND on the left, and select Subcellular location with search term "nucleus". The dropdown will offer several options but, again, do not select them. At this point, allow any assertion method for the evidence code.
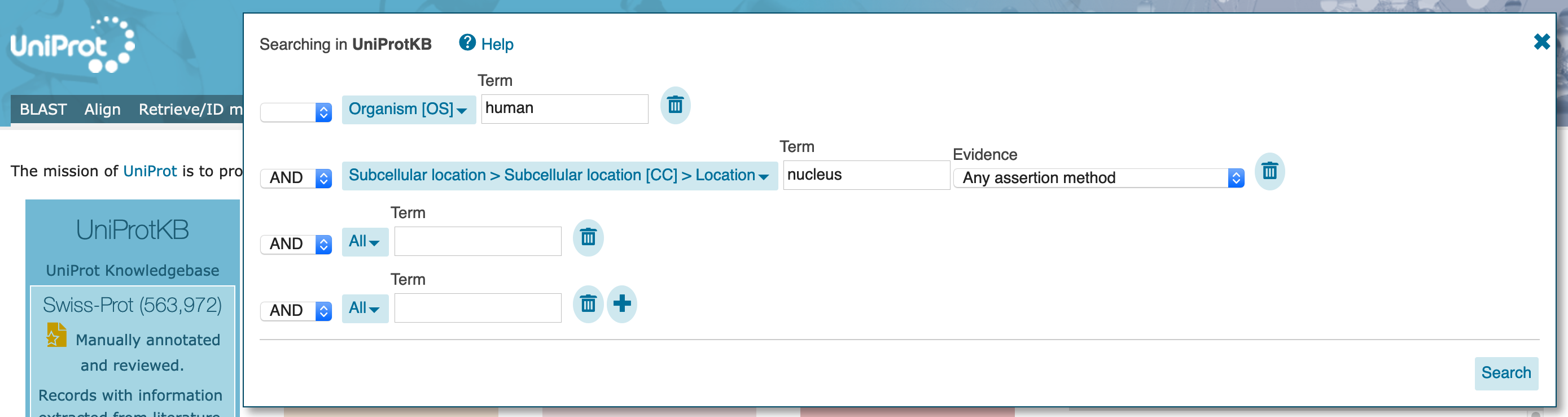
AND on the left, and select Pathology & Biotech with class "Disease", and no search term. At this point, allow any assertion method for the evidence code.
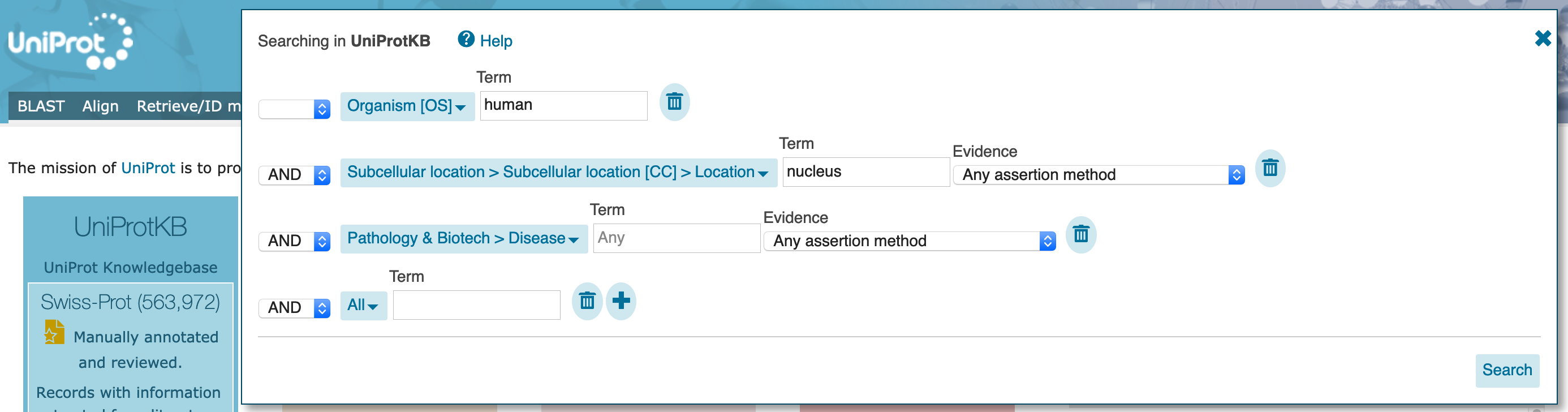
Search button.
"Advanced" drop down. You should see that the current search populates this dialogue box.
Evidence option for the `Pathology & Biotech` part of the search to manual "Experimental" evidence
Search button
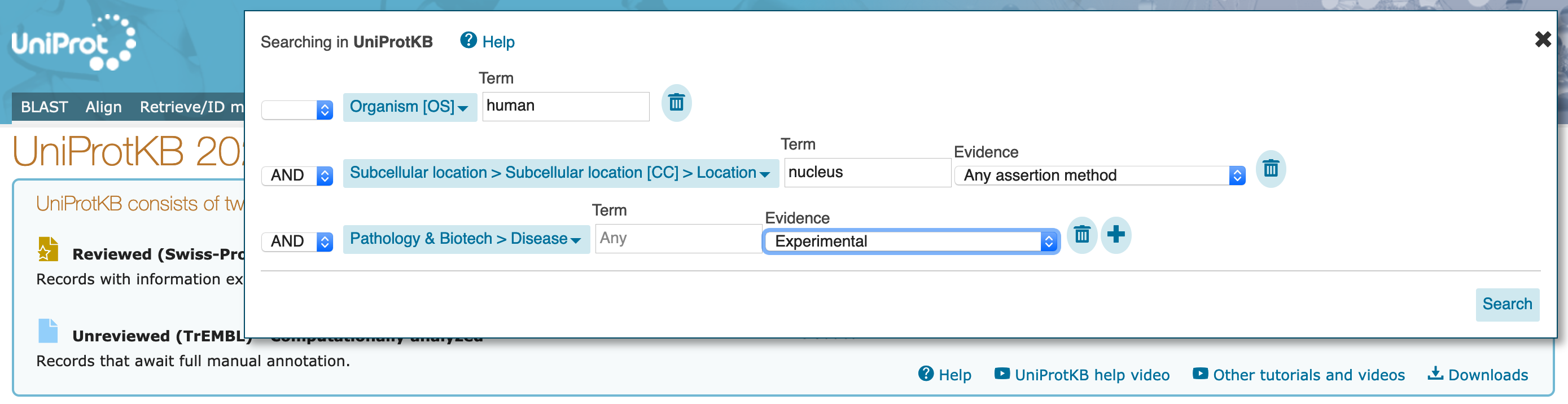
"Advanced" drop down. You should see again that the current search populates this dialogue box.
Term option for the Pathology & Biotech part of the search to "melanoma".
Search button.
UniProtKB Search Results¶After the search above, you should be left with a small set of proteins that satisfy the following criteria:
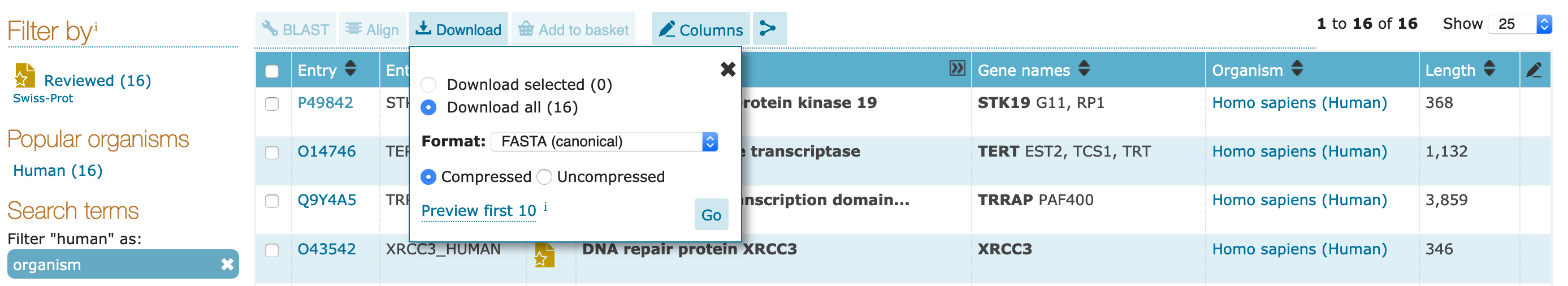
gzipped) or as raw recordsFASTA)Excel, tab-separated)XML, RDF)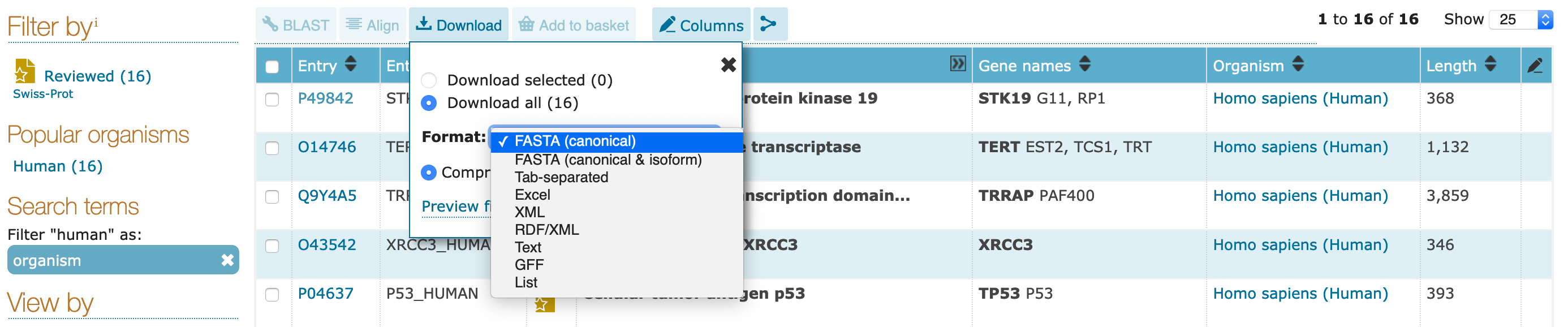

Using the UniProtKB search tools, can you find and download sets of proteins that satisfy the following requirements: