SI_Hugouvieux-Cotte-Pattat_2021
Supplementary information for pyani analyses reported in Hugouvieux-Cotte-Pattat et al. (2021) IJSEM, describing the novel genus Paradisiaca
View the Project on GitHub widdowquinn/SI_Hugouvieux-Cotte-Pattat_2021
SI_Hugovieux-Cotte-Pattat_2021
This repository contains supporting material relating to phylogenetic and pyani analyses of the Dickeya/Musicola genus, as reported in:
Hugovieux-Cotte-Pattat, N., Jacot-des-Combes, C., Briolay, J. & Pritchard, L. (2021) “Proposal for the creation of a new genus Musicola gen. nov., reclassification of Dickeya paradisiaca (Samson et al. 2005) as Musicola paradisiaca comb. nov. and description of a new species Musicola keenii sp. nov.” Int. J. Syst. Evol. Microbiol.
How to use this repository
You can use this repository like a website, to browse and see how we performed the analysis, or you can download it to inspect, verify, reproduce, or build on our analysis.
Downloading this repository
You can use git to clone this repository to your local hard drive:
git clone git@github.com:widdowquinn/SI_Hugouvieux-Cotte-Pattat_2021.git
Or you can download it as a compressed .zip archive from this link.
If You Have Problems With This Repository
Please raise an issue at the corresponding GitHub page:
Reproducing analyses (quickstart)
You can use this archive to browse, validate, reproduce, or build on the phylogenomics analysis for the Hugovieux-Cotte-Pattat et al. (2021) manuscript. We recommend creating a conda environment specific for this activity, for example using the commands:
conda create --name musicola python=3.8 -y
conda activate musicola
conda install --file requirements.txt -y
All scripts used to generate the phylogenomic analysis are found in the scripts/ subdirectory, and can be run in order to regenerate the analysis:
scripts/download_genomes.sh
scripts/annotate_genomes.sh
scripts/run_anim.sh
scripts/find_orthologues.sh
scripts/align_scos.sh
python scripts/extract_cds.py
scripts/backtranslate.sh
python scripts/concatenate_cds.py
scripts/build_tree.sh
Directory Structure
$ tree -d
.
├── figure_2 # pyani output generated for figure 2
├── figure_3 # phylogenomics support for figure 3
├── figure_S3 # pyani output generated for figure S3
├── figure_S4 # pyani output generated for figure S4
├── figures # PDF files of annotated figures from the publication
├── pyanidb # pyani database generated for figures 2,S3,S4
├── scripts # scripts used for phylogenomics and other analyses
└── sequences # FASTA sequences used to produce 16S and gapA trees
$ tree -d
.
├── figure_2 # pyani output generated for figure 2
├── figure_S3 # pyani output generated for figure S3
├── figures # PDF files of annotated figures 2 and S3
├── pyanidb # pyani database generated for figures S2/S3
└── sequences # FASTA sequences used to produce 16S and gapA trees
Reproducing Analyses
1. Whole-genome ANIm of Dickeya and Brenneria, Figures 2 and S3
The complete procedure for producing figures 2 and S3, the whole-genome ANIm of Dickeya and Brenneria spp. is described in figure2.md.
The graphical outputs relevant to figure 2 and figure S3 are presented in the directories figure_2 and figure_S3, respectively, and pyani database (readable by pyani v0.3) generated in the analysis is located in pyanidb/pyanidb_fig2_figS3.
The complete ANIm analysis, including input data, is provided in the compressed .zip archive figure_2_figure_S3.zip, which may be downloaded from DOI:10.6084/m9.figshare.14770197.
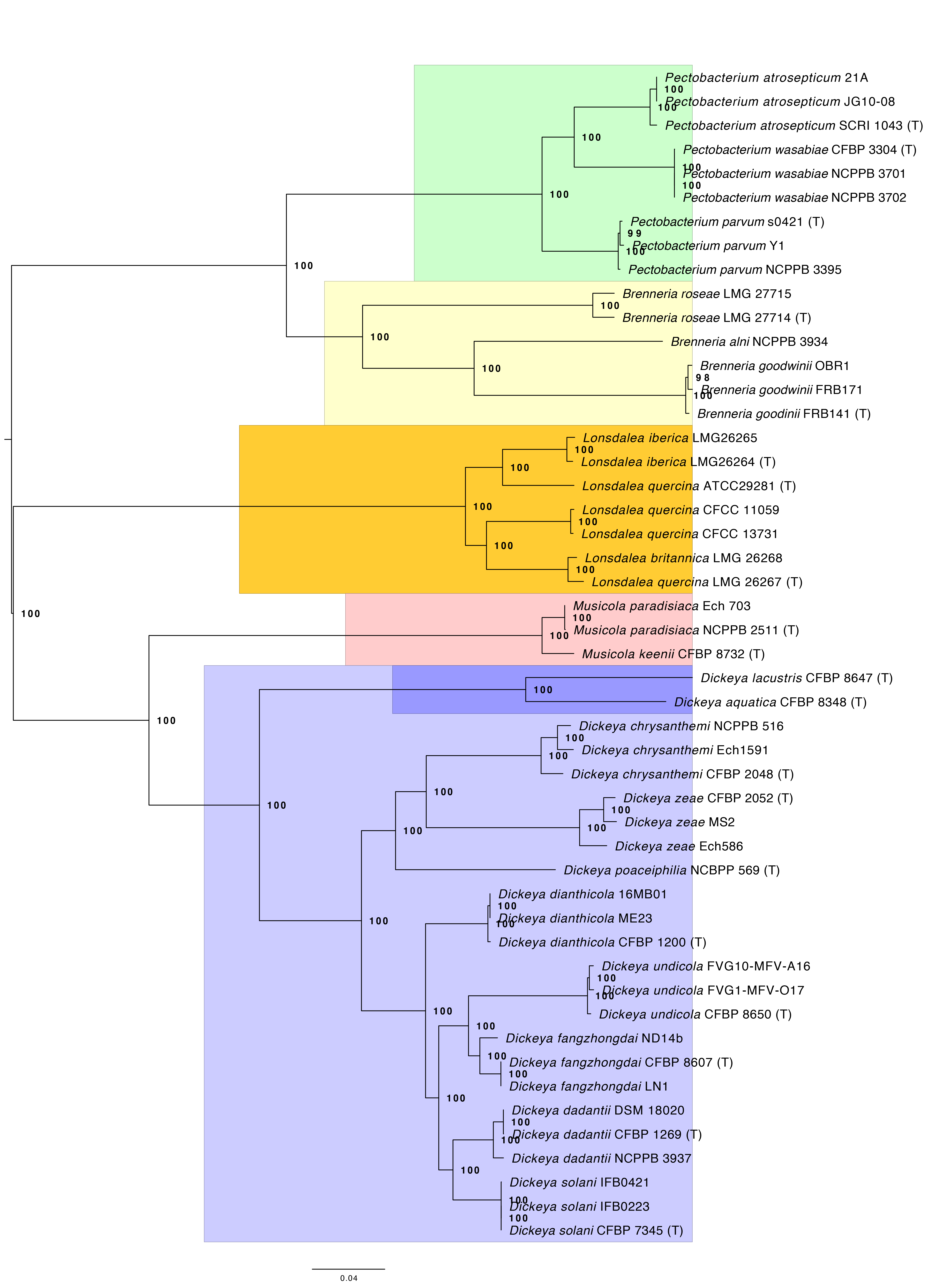
2. Phylogenomic analysis of Pectobacteriaceae, Figure 3
The complete phylogenomic analysis of 49 Pectobacteriaceae genomes, including input data, is provided in the compressed .zip archive phylogenomics.zip, which may be downloaded from 10.6084/m9.figshare.14770647. The complete procedure is outlined in the file phylogenomics_text.md. The procedure is described fully in the README.md file of the phylogenomics.zip archive, and all scripts are provided in the scripts/ subdirectory of the same archive.
Figure 3, the phylogenetic tree describing the relationship of Musicola to the other Pectobacteriaceae is provided in the figures/ directory, and corresponds to the file figures/core_genome_tree_bs_colour.pdf in the phylogenomics.zip dataset. The concatenated single-copy orthologues, partition file, raxml-ng output, log files, and FigTree output are in the figure_3/ subdirectory.
Figure S4, the ANIm analysis of Pectobacteriaceae, is split into two files: figures/figure_S4_a.pdf and figures/figure_S4_b.pdf. These correspond to the files anim_output/matrix_identity_run1.pdf and anim_output/matrix_coverage_run1.pdf from the phylogenomics.zip dataset, respectively. The complete pyani ANIm output and log files for this analysis are in the figure_S4/ subdirectory, and the pyani database (readably by pyani v0.3) is provided as the pyanidb/pyanidb_fig_S4 file.
Licensing
This repository is licensed under the MIT License, and copyright (C) 2020-2021 University of Strathclyde
If you use elements of this repository in your own work, we would be grateful if you please cite the following publication, and the URL/DOI of this repository, as “appropriate credit”.
Hugovieux-Cotte-Pattat, N., Jacot-des-Combes, C., Briolay, J. & Pritchard, L. (2021) “Proposal for the creation of a new genus Musicola gen. nov., reclassification of Dickeya paradisiaca (Samson et al. 2005) as Musicola paradisiaca comb. nov. and description of a new species Musicola keenii sp. nov.” Int. J. Syst. Evol. Microbiol.