pyani
Application and Python module for average nucleotide identity analyses of microbes.
README.md (pyani)
- README.md (pyani)
Overview
pyani is a Python3 module that provides support for calculating average nucleotide identity (ANI) and related measures for whole genome comparisons, and rendering relevant graphical summary output. Where available, it takes advantage of multicore systems, and can integrate with SGE/OGE-type job schedulers for the sequence comparisons.
pyani installs the following scripts into the $PATH:
average_nucleotide_identity.pythat enables command-line ANI analysis.genbank_get_genomes_by_taxon.pythat downloads publicly-available genomes from NCBI.delta_filter_wrapper.pyis a helper script required to run delta-filter on SGE/OGE systems.
Installation
The easiest way to install pyani v0.2 is to use conda or pip. conda is recommended for the simplest installation of third-party tool dependencies (mummer and BLAST/BLAST+).
conda
You will need to install the bioconda channel, following instructions at https://bioconda.github.io/user/install.html. Then, to create a new environment for pyani and install the program, issue the following command:
conda create --name pyani_env python=3.8 -y
conda activate pyani_env
conda install pyani
pip
pip will install pyani and its Python dependencies, but not the third-party tools.
pip3 install pyani
Docker images
pyani v0.2 scripts are also provided as Docker images, that can be run locally as containers. To use these images, first install Docker. Then, to run the corresponding scripts, issue either:
docker run -v ${PWD}:/host_dir leightonpritchard/average_nucleotide_identity
or
docker run -v ${PWD}:/host_dir leightonpritchard/genbank_get_genomes_by_taxon
NOTE
The -v ${PWD}:/host_dir argument enables the Docker container to see the current directory. Without this argument, the container will not be able to see your input files, or write output data.
Installing from the repository/source code
If you wish to install pyani v0.2 from source code, you will need to download this code from GitHub either directly, or by cloning the repository.
Both methods will create a directory called pyani which contains the source code for v0.2.
IMPORTANT NOTICE
THE DEFAULT REPOSITORY ON GITHUB IS PYANI v0.3+, NOT v0.2. THE INSTRUCTIONS ON THE MAIN REPOSITORY README.md DO NOT APPLY TO v0.2
Please note that v0.2 is a MAINTENANCE-ONLY RELEASE. This means that bugs and security issues will be fixed, but no new features will be added. All new feature development now takes place under v0.3 (the master branch on the repository).
Direct download
Download the source as .zip or .tar.gz from https://github.com/widdowquinn/pyani/releases/tag/v0.2.10
Extract the archived file using the appropriate commands (or by double-clicking in your file explorer/finder):
unzip v0.2.10.zip
or
tar -zxvf v0.2.10.tar.gz
Clone the repository using git
Download the current pyani repository with git clone, and change branch to version_0_2:
git clone git@github.com:widdowquinn/pyani.git
git checkout version_0_2
Installation
Change directory to pyani, and use the python setup.py setuptools command, to install the package and scripts. This will not install the third-party tools BLAST/BLAST+ and mummer.
cd pyani
python setup.py build
python setup.py install
Third-party tools
Three alignment packages are required, to use all of pyani’s methods: mummer, BLAST+, and legacy BLAST.
The simplest route to obtaining these tools is to use conda/bioconda:
conda install mummer blast legacy-blast -y
But they can also be installed by following instructions from the tools’ own websites.
- BLAST+ (for
anib) ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/ - legacy BLAST (for
aniblastall) ftp://ftp.ncbi.nlm.nih.gov/blast/executables/release/LATEST/ - MUMmer (for
anim) http://mummer.sourceforge.net/Testing
pyani
The pyani repository includes tests that can be run with pytest (including coverage measurement using pytest-cov) with the following command, executed from the repository root directory:
pytest -v
Running pyani
Script: average_nucleotide_identity.py
The average_nucleotide_identity.py script - installed as part of this package - enables straightforward ANI analysis at the command-line, and uses the pyani module behind the scenes.
You can get a summary of available command-line options with average_nucleotide_identity.py -h
$ average_nucleotide_identity.py -h
usage: average_nucleotide_identity.py [-h] [-o OUTDIRNAME] [-i INDIRNAME] [-v]
[-f] [-s FRAGSIZE] [-l LOGFILE]
[--skip_nucmer] [--skip_blastn]
[--noclobber] [--nocompress] [-g]
[--gformat GFORMAT] [--gmethod GMETHOD]
[--labels LABELS] [--classes CLASSES]
[-m METHOD] [--scheduler SCHEDULER]
[--workers WORKERS]
[--SGEgroupsize SGEGROUPSIZE]
[--maxmatch] [--nucmer_exe NUCMER_EXE]
[--blastn_exe BLASTN_EXE]
[--makeblastdb_exe MAKEBLASTDB_EXE]
[--blastall_exe BLASTALL_EXE]
[--formatdb_exe FORMATDB_EXE]
[--write_excel] [--subsample SUBSAMPLE]
[--seed SEED] [--jobprefix JOBPREFIX]
[…]
Example data and output can be found in the directory test_ani_data. The data are chromosomes of four isolates of Caulobacter. Basic analyses can be performed with the command lines:
average_nucleotide_identity.py -i tests/test_ani_data/ -o tests/test_ANIm_output -m ANIm -g
average_nucleotide_identity.py -i tests/test_ani_data/ -o tests/test_ANIb_output -m ANIb -g
average_nucleotide_identity.py -i tests/test_ani_data/ -o tests/test_ANIblastall_output -m ANIblastall -g
average_nucleotide_identity.py -i tests/test_ani_data/ -o tests/test_TETRA_output -m TETRA -g
The graphical output below, supporting assignment of NC_002696 and NC_011916 to the same species (C.crescentus), and the other two isolates to distinct species (NC_014100:C.segnis; NC_010338:C. sp K31), was generated with the command-line:
average_nucleotide_identity.py -v -i tests/test_ani_data/ \
-o tests/test_ANIm_output/ -g --gformat png,pdf,eps \
--classes tests/test_ani_data/classes.tab \
--labels tests/test_ani_data/labels.tab
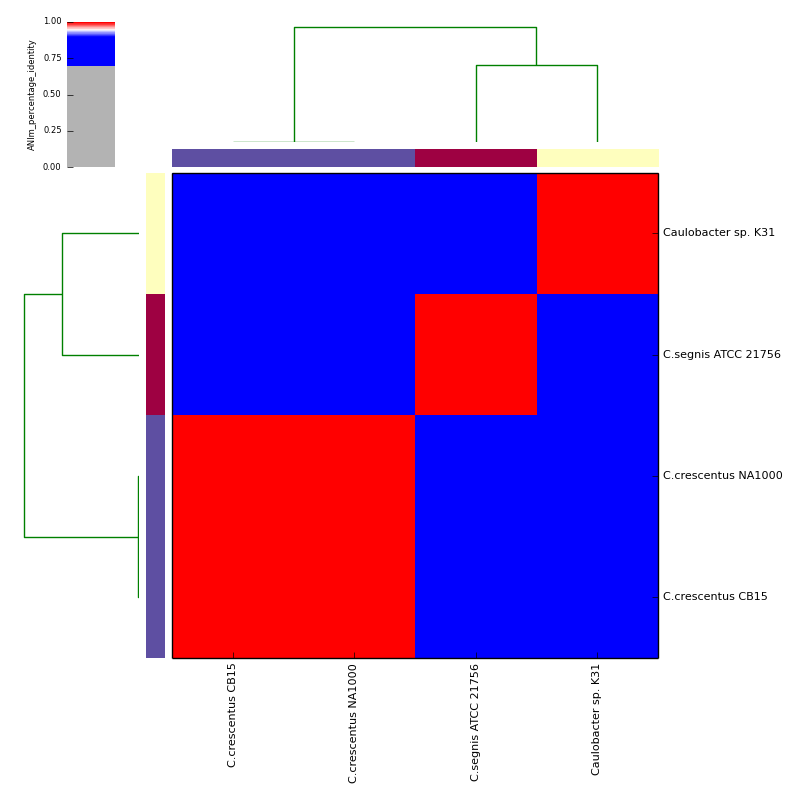
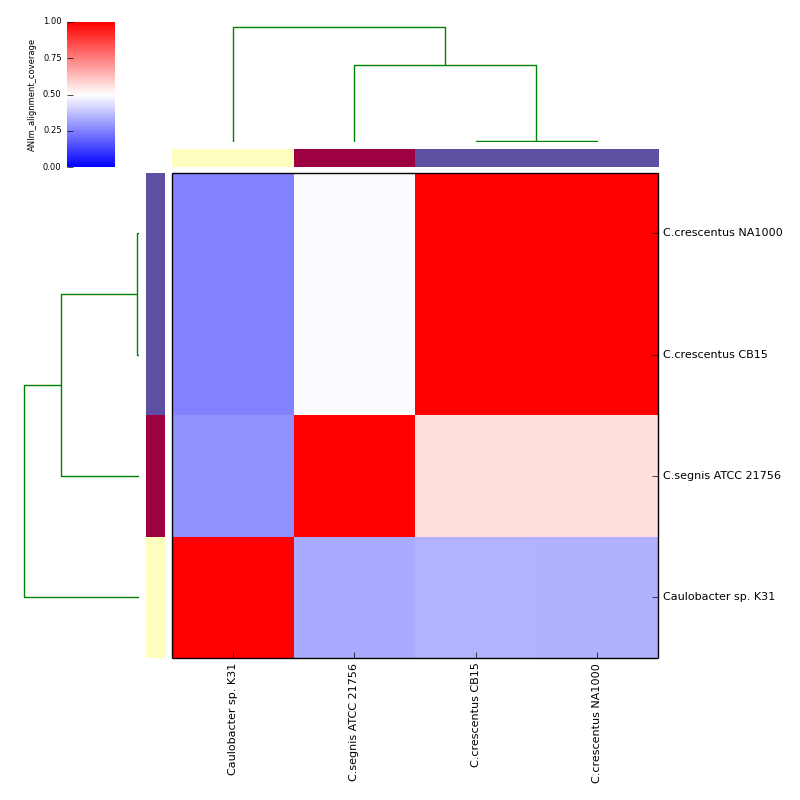
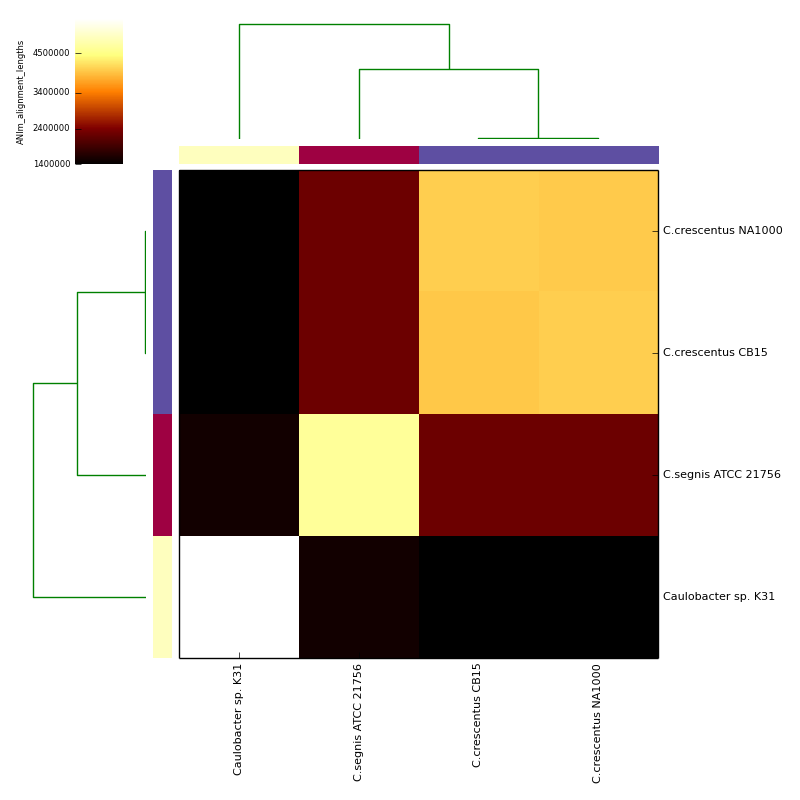
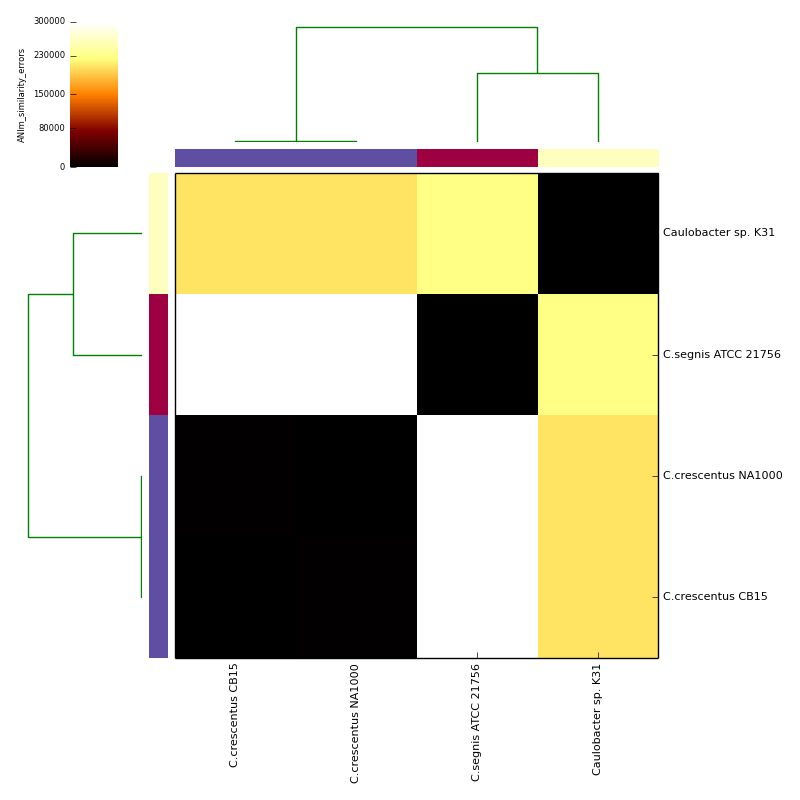
Script: genbank_get_genomes_by_taxon.py
The script genbank_get_genomes_by_taxon.py, installed by this package, enables download of genomes from NCBI, specified by taxon ID. The script will download all available assemblies for taxa at or below the specified node in the NCBI taxonomy tree.
Command-line options can be viewed using:
$ genbank_get_genomes_by_taxon.py -h
usage: genbank_get_genomes_by_taxon.py [-h] [-o OUTDIRNAME] [-t TAXON] [-v]
[-f] [--noclobber] [-l LOGFILE]
[--format FORMAT] [--email EMAIL]
[--retries RETRIES]
[--batchsize BATCHSIZE]
[…]
For example, the NCBI taxonomy ID for Caulobacter is 75, so all publicly-available Caulobacter sequences can be obtained using the command-line:
$ genbank_get_genomes_by_taxon.py -o Caulobacter_downloads -v -t 75 -l Caulobacter_downloads.log --email me@my.email.domain
INFO: genbank_get_genomes_by_taxon.py: Mon Apr 18 17:22:54 2016
INFO: command-line: /Users/lpritc/Virtualenvs/pyani3/bin/genbank_get_genomes_by_taxon.py -o Caulobacter_downloads -v -t 75 -l Caulobacter_downloads.log --email me@my.email.domain
INFO: Namespace(batchsize=10000, email='me@my.email.domain', force=False, format='gbk,fasta', logfile='Caulobacter_downloads.log', noclobber=False, outdirname='Caulobacter_downloads', retries=20, taxon='75', verbose=True)
INFO: Set NCBI contact email to me@my.email.domain
INFO: Creating directory Caulobacter_downloads
INFO: Output directory: Caulobacter_downloads
INFO: Passed taxon IDs: 75
INFO: Entrez ESearch with query: txid75[Organism:exp]
INFO: Entrez ESearch returns 29 assembly IDs
INFO: Identified 29 unique assemblies
INFO: Taxon 75: 29 assemblies
[…]
INFO: Assembly 639581: 271 contigs
INFO: Assembly 233261: 17 contigs
INFO: Assembly 575291: 48 contigs
INFO: Mon Apr 18 17:25:46 2016
INFO: Done.
NOTE: You must provide a valid email to identify yourself to NCBI for troubleshooting.
The number of attempted retries for each download, and the size of a batch download can be modified. By default, the script will attempt 20 download retries, and obtain sequences in batches of 10000.
Method and Output Description
Average Nucleotide Identity (ANI)
This module calculates Average Nucleotide Identity (ANI) according to one of a number of alternative methods described in, e.g.
- Richter M, Rossello-Mora R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106: 19126-19131. doi:10.1073/pnas.0906412106. (ANI1020, ANIm, ANIb)
- Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, et al. (2007) DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Micr 57: 81-91. doi:10.1099/ijs.0.64483-0.
ANI is proposed to be the appropriate in silico substitute for DNA-DNA hybridisation (DDH), and so useful for delineating species boundaries. A typical percentage threshold for species boundary in the literature is 95% ANI (e.g. Richter et al. 2009).
All ANI methods follow the basic algorithm:
- Align the genome of organism 1 against that of organism 2, and identify the matching regions
- Calculate the percentage nucleotide identity of the matching regions, as an average for all matching regions
Methods differ on: (1) what alignment algorithm is used, and the choice of parameters (this affects the aligned region boundaries); (2) what the input is for alignment (typically either fragments of fixed size, or the most complete assembly available).
- ANIm: uses MUMmer (NUCmer) to align the input sequences.
- ANIb: uses BLASTN+ to align 1020nt fragments of the input sequences
- ANIblastall: uses legacy BLASTN to align 1020nt fragments of the input sequences
- TETRA: calculates tetranucleotide frequencies of each input sequence
The algorithms takes as input correctly-formatted FASTA multiple sequence files. All sequences for a single organism should be contained in only one sequence file. Although it is possible to provide new labels for each input genome, for rendering graphical output, the names of these files are used for identification so it is best to name them sensibly.
Output is written to a named directory. The output files differ depending on the chosen ANI method.
- ANIm: MUMmer/NUCmer .delta files, describing each pairwise sequence alignment. Output as tab-separated plain text format tables describing: alignment coverage; total alignment lengths; similarity errors; and percentage identity (ANIm).
- ANIb and ANIblastall: FASTA sequences describing 1020nt fragments of each input sequence; BLAST nucleotide databases - one for each set of fragments; and BLASTN output files (tab-separated tabular format plain text) - one for each pairwise comparison of input sequences. Output as tab-separated plain text tables describing: alignment coverage; total alignment lengths; similarity errors; and percentage identity (ANIb or ANIblastall).
- TETRA: Tab-separated plain text files describing the Pearson correlations between Z-score distributions for each tetranucleotide in each input sequence (TETRA).
If graphical output is chosen, the output directory will also contain PDF, PNG and EPS files representing the various output measures as a heatmap with row and column dendrograms. Other output formats (e.g. SVG) can be specified with the --gformat argument.
Licensing
Unless otherwise indicated, all code is subject to the following agreement:
(c) The James Hutton Institute 2014-2019 (c) The University of Strathclyde 2019-2021 Author: Leighton Pritchard
Contact: leighton.pritchard@strath.ac.uk
Address: Leighton Pritchard Strathclyde Institute for Pharmacy and Biomedical Sciences University of Strathclyde 161 Cathedral Street Glasgow Scotland G4 0RE UK
The MIT License
Copyright (c) 2014-2019 The James Hutton Institute Copyright (c) 2019-2021 The University of Strathclyde
Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the “Software”), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED “AS IS”, WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.



